
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,865,342 – 10,865,435 |
| Length | 93 |
| Max. P | 0.633717 |

| Location | 10,865,342 – 10,865,435 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.56 |
| Shannon entropy | 0.45130 |
| G+C content | 0.59823 |
| Mean single sequence MFE | -38.15 |
| Consensus MFE | -23.10 |
| Energy contribution | -22.41 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.633717 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10865342 93 + 21146708 AUAGCAGCCUUGCCAACGGAG---------------AUGCCUACUACUGCACGGGCAGUAACCAUGAGUGCUGCUCGGGCA------------ACGAUGUGGUGCUCAUUAAGGUGGGCA ...((......))........---------------.(((((((((((((....)))))....(((((..(..((((....------------.))).)..)..)))))...)))))))) ( -35.50, z-score = -0.97, R) >droGri2.scaffold_15245 2938530 102 + 18325388 GCGGUGGCCUGAGCCAUGUCGGUGGGG------GGGAUGCCUAUUAUUGCACGGGCAGCAAUCACGAGUGUUGCAACGGCA------------ACGAUGUGGUGCUCAUUAAGGUGGGUG .((.(.((((..(((.(((((((((.(------((....))).)))))).)))))).........(((..(..((.((...------------.)).))..)..)))....)))).).)) ( -36.20, z-score = -0.33, R) >droMoj3.scaffold_6496 9523321 102 - 26866924 GAAGUGGCCUGGGCACUGCGGGUGGGG------GCGAUGCCUACUACUGCACGGGCAGCAAUCACGAGUGCUGCAACGGCA------------ACGAUGUGGUGCUCAUUAAGGUGGGUG ...((.((((...(((.....))).))------)).))(((((((.((((....)))).......(((..(..((.((...------------.)).))..)..))).....))))))). ( -41.10, z-score = -1.25, R) >droVir3.scaffold_12875 9730286 102 + 20611582 GCGGUGGCCUAAGCACCGCCAGUGGGG------GGGAUGCCUACUACUGCACGGGCAGCAAUCACGAGUGCUGUAACGGCA------------ACGAUGUGGUGCUCAUUAAGGUGGGUG .((.(.((((..((.(((.((((((..------((....))..))))))..))))).........(((..(..((.((...------------.)).))..)..)))....)))).).)) ( -40.80, z-score = -0.86, R) >droWil1.scaffold_180699 1135541 99 - 2593675 -------UCUGGUCAAGGAUGGCG------------GUGC-UAUUACCGUGCUGGCGACAAUCAUGAGUGCUGCACGGACACGGGCACGGGCGACGAUGUGGUGCUCAUUAAGGUGGGG- -------....((((....)))).------------((((-(....((((((.(((.((........))))))))))).....)))))((((.((......))))))............- ( -30.10, z-score = 0.78, R) >droAna3.scaffold_13266 17424480 108 - 19884421 ACAGCAGUAUCAGCAGCGGAGGAGGAGGAGGAGGAGACGCCUACUACUGCACGGGCAGCAAUCAUGAGUGCUGCUCCGGCA------------ACGAUGUGGUGCUCAUUAAGGUGGGCA ...((..((((.((.((((...((....(((.(....).))).)).)))).(((((((((........)))))).))))).------------..))))..))((((((....)))))). ( -39.00, z-score = -1.07, R) >droEre2.scaffold_4845 7600867 93 - 22589142 ACAGCAGCCUCGCCAACGGAG---------------ACGCCUACUACUGCACGGGCAGCAACCACGAGUGUUGCUCGGGCA------------ACGACGUGGUGCUCAUUAAGGUGGGCA ......(((.((((...(...---------------.)((((..........))))((((.(((((..((((((....)))------------))).)))))))))......))))))). ( -38.30, z-score = -1.64, R) >droYak2.chr2R 10795833 93 + 21139217 ACAGCAGCCUCACCAACGGAG---------------ACGCCUACUACUGCACGGGCAGCAACCACGAGUGUUGCUCGGGCA------------ACGACGUGGUGCUCAUUAAGGUGGGCA ......(((.((((...(...---------------.)((((..........))))((((.(((((..((((((....)))------------))).)))))))))......))))))). ( -38.70, z-score = -2.24, R) >droSec1.super_1 8377926 93 + 14215200 ACAGCAGCCUCGCCAACGGAG---------------ACGCCUACUACUGCACGGGCAGCAACCACGAGUGCUGCUCGGGCA------------ACGAUGUGGUGCUCAUUAAGGUGGGCA ......(((.((((....(((---------------.((((.((...(((.(((((((((........))))))))).)))------------.....))))))))).....))))))). ( -40.90, z-score = -2.30, R) >droSim1.chr2R 9625756 93 + 19596830 ACAGCAGCCUCGCCAACGGAG---------------ACGCCUACUACUGCACGGGCAGCAACCAUGAGUGCUGCUCGGGCA------------ACGAUGUGGUGCUCAUUAAGGUGGGCA ......(((.((((....(((---------------.((((.((...(((.(((((((((........))))))))).)))------------.....))))))))).....))))))). ( -40.90, z-score = -2.30, R) >consensus ACAGCAGCCUCGCCAACGGAG_______________ACGCCUACUACUGCACGGGCAGCAACCACGAGUGCUGCUCGGGCA____________ACGAUGUGGUGCUCAUUAAGGUGGGCA ......................................(((((((.((((....)))).......(((..(..(........................)..)..))).....))))))). (-23.10 = -22.41 + -0.69)

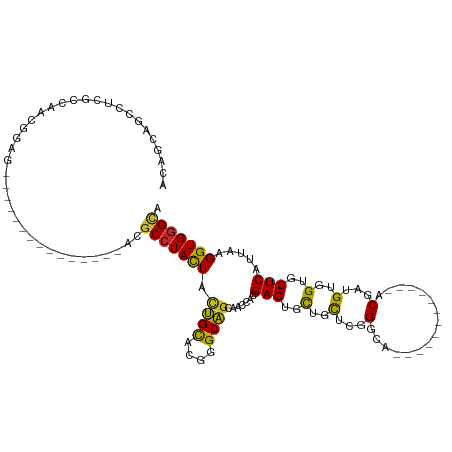
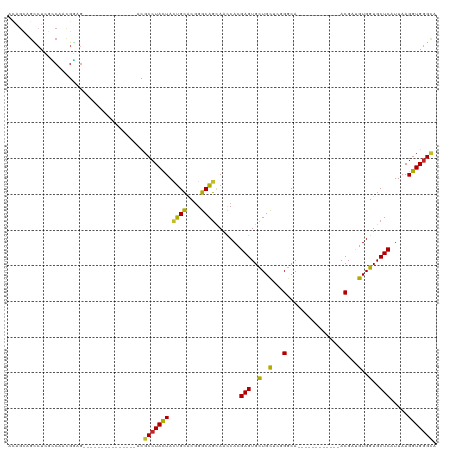
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:23:51 2011