
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,859,535 – 10,859,640 |
| Length | 105 |
| Max. P | 0.793831 |

| Location | 10,859,535 – 10,859,640 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.97 |
| Shannon entropy | 0.49431 |
| G+C content | 0.62420 |
| Mean single sequence MFE | -45.03 |
| Consensus MFE | -28.30 |
| Energy contribution | -26.65 |
| Covariance contribution | -1.66 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.793831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
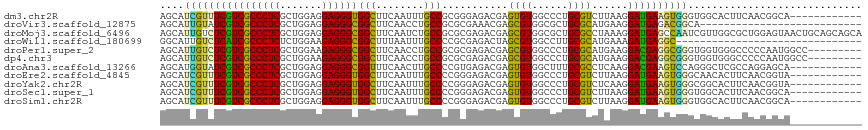
>dm3.chr2R 10859535 105 + 21146708 AGCAUCGUUUCGUCGCCCUCGCUGGAGGAGGGUGGCUUCAAUUUGCCGCGGGAGACGAGUGUGGCCCUGCGUCUUAAGGAUGAAGUGGGUGGCACUUCAACGGCA------------ ...........(((((((((.......))))))))).......(((((..(....)((((((.((((..((((.....))))....)))).))))))...)))))------------ ( -44.70, z-score = -1.55, R) >droVir3.scaffold_12875 9719938 90 + 20611582 AGCAUUGUAUCGUCGCCCUCGCUGGAGGAGGGCGGCUUCAACCUGCCGCGCGAAACGAGCGUGGCGCUGCGCAUGAAGGAUGAGACGGCA--------------------------- .((........(((((((((.......)))))))))((((.((((((((((.......)))))))((...))....))).))))...)).--------------------------- ( -40.30, z-score = -1.16, R) >droMoj3.scaffold_6496 9513783 117 - 26866924 AGCAUUGUCUCGUCGCCCUCGCUGGAGGAGGGCGGCUUCAAUCUGCCGCGCGAGACGAGCGUGGCGCUGCGCCUAAAGGAUGAGCCAAUCGUUGGCGCUGGAGUAACUGCAGCAGCA ((.((((....(((((((((.......)))))))))..))))))(((((((.......)))))))((((((((..((.(((......))).)))))((.(......).)).))))). ( -52.80, z-score = -0.63, R) >droWil1.scaffold_180699 1127561 86 - 2593675 GGCAUUGUCUCAUCGCCCUCUCUGGAAGAGGGCGGCUUUAAUUUGCCCCGCGAGACUAGCGUGGCCUUGCGCAUGAAAGAUGAGGC------------------------------- ......(((((((((((((((.....)))))))(((........))).((((((.((.....)).)))))).......))))))))------------------------------- ( -36.30, z-score = -2.19, R) >droPer1.super_2 759883 108 - 9036312 AGCAUUGUCUCGUCGCCCUCGCUGGAAGAGGGCGGCUUCAACCUGCCGCGCGAGACGAGCGUGGCCCUGCGCAUGAAGGACGAGGCGGGUGGUGGGCCCCCAAUGGCC--------- ..((((((((((((((((((.......))))))..(((((.((.(((((((.......)))))))...).)..)))))))))))))(((.((....)))))))))...--------- ( -52.90, z-score = -1.08, R) >dp4.chr3 13345498 108 + 19779522 AGCAUUGUCUCGUCGCCCUCGCUGGAAGAGGGCGGCUUCAACCUGCCGCGCGAGACGAGCGUGGCCCUGCGCAUGAAGGACGAGGCGGGUGGUGGGCCCCCAAUGGCC--------- ..((((((((((((((((((.......))))))..(((((.((.(((((((.......)))))))...).)..)))))))))))))(((.((....)))))))))...--------- ( -52.90, z-score = -1.08, R) >droAna3.scaffold_13266 17419274 105 - 19884421 AGCAUGGUAUCGUCGCCCUCGCUGGAGGAGGGCGGUUUCAACUUGCCCCGUGAGACGAGUGUGGCUUUGCGCCUCAAGGACGAAGUCCAGGGCUCGCCAGGAGCA------------ .((.((((....((((((((.......)))))))).........(((((....)..(((.(((......))))))..((((...)))).))))..))))...)).------------ ( -39.40, z-score = 0.65, R) >droEre2.scaffold_4845 7595009 105 - 22589142 AGCAUCGUUUCGUCGCCCUCGCUGGAGGAGGGUGGCUUCAAUUUGCCCCGGGAGACGAGUGUGGCCCUGCGUCUUAAGGAUGAAGUGGGCAACACUUCAACGGUA------------ .(((.......(((((((((.......))))))))).......))).((((....)((((((.((((..((((.....))))....)))).))))))...)))..------------ ( -42.54, z-score = -1.48, R) >droYak2.chr2R 10789812 105 + 21139217 AGCAUCGUUUCGUCGCCCUCGCUGGAGGAGGGUGGCUUCAAUUUGCCCCGGGAGACGAGUGUGGCCCUGCGUCUCAAGGAUGAAGUGGGCGGCACUUCAACGGUA------------ .(((.......(((((((((.......))))))))).......))).((((....)((((((.((((..((((.....))))....)))).))))))...)))..------------ ( -44.94, z-score = -1.32, R) >droSec1.super_1 8372054 105 + 14215200 AGCAUCGUUUCGUCGCCCUCGCUGGAGGAGGGUGGCUUCAAUUUGCCCCGGGAGACGAGUGUGGCCCUGCGUCUUAAGGAUGAAGUGGGUGGCACUUCAACGGCA------------ .((.((((((((((((((((.......))))))))).........(....))))))))((((.((((..((((.....))))....)))).)))).......)).------------ ( -44.30, z-score = -1.34, R) >droSim1.chr2R 9619891 105 + 19596830 AGCAUCGUUUCGUCGCCCUCGCUGGAGGAGGGUGGCUUCAAUUUGCCCCGGGAGACGAGUGUGGCCCUGCGUCUUAAGGAUGAAGUGGGUGGCACUUCAACGGCA------------ .((.((((((((((((((((.......))))))))).........(....))))))))((((.((((..((((.....))))....)))).)))).......)).------------ ( -44.30, z-score = -1.34, R) >consensus AGCAUCGUUUCGUCGCCCUCGCUGGAGGAGGGCGGCUUCAAUUUGCCCCGCGAGACGAGUGUGGCCCUGCGCCUGAAGGAUGAAGUGGGUGGCACUUCAACGGUA____________ ....((((((((((((((((.......))))))(((........)))...........((((......))))......))))))))))............................. (-28.30 = -26.65 + -1.66)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:23:50 2011