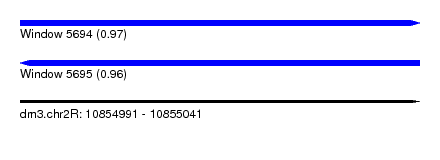
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,854,991 – 10,855,041 |
| Length | 50 |
| Max. P | 0.968655 |

| Location | 10,854,991 – 10,855,041 |
|---|---|
| Length | 50 |
| Sequences | 7 |
| Columns | 61 |
| Reading direction | forward |
| Mean pairwise identity | 64.73 |
| Shannon entropy | 0.58804 |
| G+C content | 0.44829 |
| Mean single sequence MFE | -13.23 |
| Consensus MFE | -9.95 |
| Energy contribution | -9.10 |
| Covariance contribution | -0.85 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
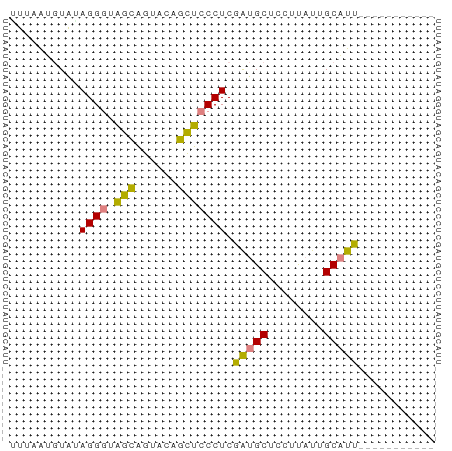
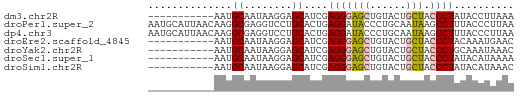
>dm3.chr2R 10854991 50 + 21146708 UUUAAGGUAUAGGGUAGCAGUACAGCUCCCUCGAUGCUCCUUAUUGCAUU----------- ..........((((.(((......))))))).(((((........)))))----------- ( -12.80, z-score = -1.17, R) >droPer1.super_2 755926 61 - 9036312 UUAAGGGUAAAGGCUUAUUGCAGGGUAUCCUCAGUGCAGGACCUCGCCUUGUUAAUGCAUU ............((.....(((((((.((((......))))....)))))))....))... ( -13.80, z-score = 0.41, R) >dp4.chr3 13341505 61 + 19779522 UUAAGGGUAAAGGCUUAUUGCAGGGUAUCCUCAGUGCAGGACCUCGCCUUGUUAAUGCAUU ............((.....(((((((.((((......))))....)))))))....))... ( -13.80, z-score = 0.41, R) >droEre2.scaffold_4845 7590469 50 - 22589142 GUUCAUUUGUAGGGUAGCAGUACAGCUCCCUCGAUGCUCCUUAUUGCAUU----------- ........(.((((.(((......))))))))(((((........)))))----------- ( -13.30, z-score = -1.95, R) >droYak2.chr2R 10785174 50 + 21139217 GUUUAUUUGCAGGGUAGCAGUACAGCUCCCUCGAUGCUCCUUAUUGCAUU----------- ........(.((((.(((......))))))))(((((........)))))----------- ( -13.30, z-score = -1.63, R) >droSec1.super_1 8367570 50 + 14215200 UUUUAUGUAUAGGGUAGCAGUACAGCUCCCUCGAUGCUCCUUAUUGCAUU----------- ..........((((.(((......))))))).(((((........)))))----------- ( -12.80, z-score = -1.71, R) >droSim1.chr2R 9615367 50 + 19596830 GUUUAUGUAUAGGGUAGCAGUACAGCUCCCUCGAUGCUCCUUAUUGCAUU----------- ..........((((.(((......))))))).(((((........)))))----------- ( -12.80, z-score = -1.47, R) >consensus UUUAAUGUAUAGGGUAGCAGUACAGCUCCCUCGAUGCUCCUUAUUGCAUU___________ ..........((((.(((......))))))).(((((........)))))........... ( -9.95 = -9.10 + -0.85)

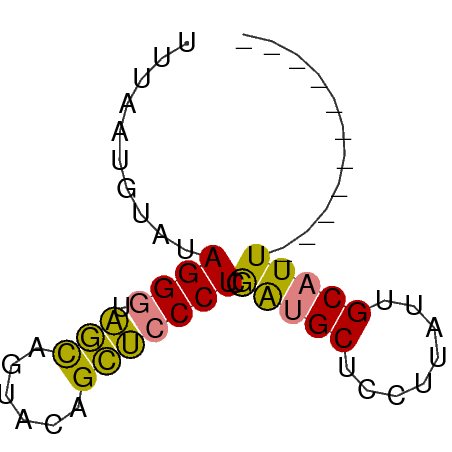
| Location | 10,854,991 – 10,855,041 |
|---|---|
| Length | 50 |
| Sequences | 7 |
| Columns | 61 |
| Reading direction | reverse |
| Mean pairwise identity | 64.73 |
| Shannon entropy | 0.58804 |
| G+C content | 0.44829 |
| Mean single sequence MFE | -12.13 |
| Consensus MFE | -7.56 |
| Energy contribution | -7.31 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.959925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10854991 50 - 21146708 -----------AAUGCAAUAAGGAGCAUCGAGGGAGCUGUACUGCUACCCUAUACCUUAAA -----------.......(((((.......(((((((......))).))))...))))).. ( -13.50, z-score = -1.81, R) >droPer1.super_2 755926 61 + 9036312 AAUGCAUUAACAAGGCGAGGUCCUGCACUGAGGAUACCCUGCAAUAAGCCUUUACCCUUAA ...........((((.(((((..((((....((....))))))....)))))...)))).. ( -10.90, z-score = 0.21, R) >dp4.chr3 13341505 61 - 19779522 AAUGCAUUAACAAGGCGAGGUCCUGCACUGAGGAUACCCUGCAAUAAGCCUUUACCCUUAA ...........((((.(((((..((((....((....))))))....)))))...)))).. ( -10.90, z-score = 0.21, R) >droEre2.scaffold_4845 7590469 50 + 22589142 -----------AAUGCAAUAAGGAGCAUCGAGGGAGCUGUACUGCUACCCUACAAAUGAAC -----------.((((........))))..(((((((......))).)))).......... ( -12.40, z-score = -1.97, R) >droYak2.chr2R 10785174 50 - 21139217 -----------AAUGCAAUAAGGAGCAUCGAGGGAGCUGUACUGCUACCCUGCAAAUAAAC -----------.((((........))))..(((((((......))).)))).......... ( -12.40, z-score = -1.53, R) >droSec1.super_1 8367570 50 - 14215200 -----------AAUGCAAUAAGGAGCAUCGAGGGAGCUGUACUGCUACCCUAUACAUAAAA -----------.((((........))))..(((((((......))).)))).......... ( -12.40, z-score = -1.75, R) >droSim1.chr2R 9615367 50 - 19596830 -----------AAUGCAAUAAGGAGCAUCGAGGGAGCUGUACUGCUACCCUAUACAUAAAC -----------.((((........))))..(((((((......))).)))).......... ( -12.40, z-score = -1.93, R) >consensus ___________AAUGCAAUAAGGAGCAUCGAGGGAGCUGUACUGCUACCCUAUACAUAAAA ..............((........))....(((((((......))).)))).......... ( -7.56 = -7.31 + -0.24)
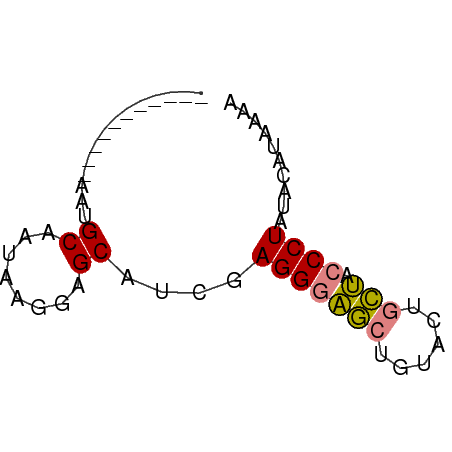
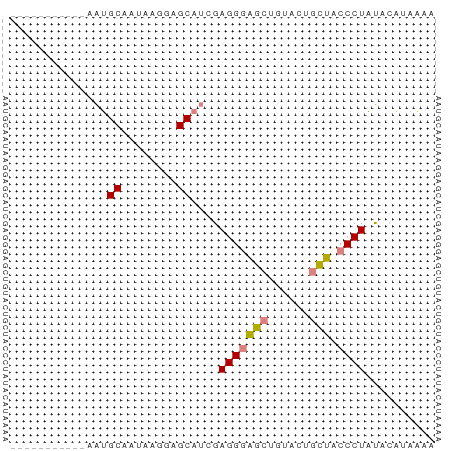
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:23:46 2011