
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,794,278 – 10,794,350 |
| Length | 72 |
| Max. P | 0.582494 |

| Location | 10,794,278 – 10,794,350 |
|---|---|
| Length | 72 |
| Sequences | 9 |
| Columns | 74 |
| Reading direction | reverse |
| Mean pairwise identity | 60.96 |
| Shannon entropy | 0.81723 |
| G+C content | 0.38248 |
| Mean single sequence MFE | -10.93 |
| Consensus MFE | -4.60 |
| Energy contribution | -4.50 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.48 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.582494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10794278 72 - 21146708 -ACGCUCAGCCAAUUUGGCACCGCUAUGAAAAAACAAUCACUG-AAAAAUAUUUUGCCAGUUUUAAUGGUGUUA -.......(((.....)))..((((((((........))((((-.(((....)))..))))....))))))... ( -9.90, z-score = 0.71, R) >droSim1.chr2R 9554395 73 - 19596830 -ACGCUUAGCCAAUUUGGCACCGCUAUGAAAAAACAAUCACUGAAAAAAUAUUUAGACAGUUUUAAUGGUAUUG -.......(((.....)))...(((((...(((((.....(((((......)))))...))))).))))).... ( -10.80, z-score = -0.61, R) >droSec1.super_1 8307808 72 - 14215200 -ACGCUUAGCCAGUUUGGCACCGCUAUGAAAAAACAAUCACUG-AAAAAUAUUUAGCCAGUUUUAAUGGUAUUG -.......(((.....)))...(((((...(((((.....(((-((.....)))))...))))).))))).... ( -10.60, z-score = 0.21, R) >droEre2.scaffold_4845 7530810 65 + 22589142 -ACGCUUAGCCAAUUUGGCACCGCCAAGAAAA------CACU--GAAAAUAUUUACGCUCUACAAAAGGUGUUA -..((...(((.....)))...))......((------((((--.......................)))))). ( -8.80, z-score = 0.37, R) >droVir3.scaffold_12875 9637043 65 - 20611582 ACAGCUCAGCCAACUUGGCACCG---ACAGCACACAAAAAAU--AGAAAUC-GUUU-CGGUU--AUUUUUGCUA ..(((...(((.....)))((((---(.(((...........--.......-))))-)))).--......))). ( -12.17, z-score = -1.23, R) >droGri2.scaffold_15245 2847156 61 - 18325388 ACAGCGCAGUCAACUUGGCACCG--------ACACAAAAAAU--AGAAAUCUUUCU-GCUUU--AUUUUUGUUA ...((.(((.....)))))...(--------(((.(((((((--((((....))))-).)))--.))).)))). ( -6.00, z-score = 0.82, R) >droMoj3.scaffold_6496 9408105 61 + 26866924 ACAGCUGAGCCAACUUGGCAGCG--CACAGCGCGCACAAAAU--AGAAAUC-UUCU-CUGCU--AUUUU----- ..(((.(((((.....))).(((--(.....)))).......--.......-...)-).)))--.....----- ( -15.60, z-score = -0.98, R) >droWil1.scaffold_180700 3659451 66 + 6630534 -GCUCUCAGCCAACUUGGCACUG---UCAAUUGGCAAAAAAU--AGAAAUGUUUGU-CGUUUUCGUUUUUGUU- -.......(((((.(((((...)---))))))))).((((((--..(((((.....-)))))..))))))...- ( -16.20, z-score = -2.17, R) >droYak2.chr2R 10723858 72 - 21139217 -ACGCUCAGCCAAUUUGGCACCGCUAAGAAAAAACAAACACUG-AAAAAUACGAGUAUUAACACACUAUAAUAG -..((((.(((.....)))...............((.....))-........)))).................. ( -8.30, z-score = -1.47, R) >consensus _ACGCUCAGCCAAUUUGGCACCGC_AAGAAAAAACAAACACU__AAAAAUAUUUAU_CAGUUU_AAUGUUGUUA ........(((.....)))....................................................... ( -4.60 = -4.50 + -0.10)

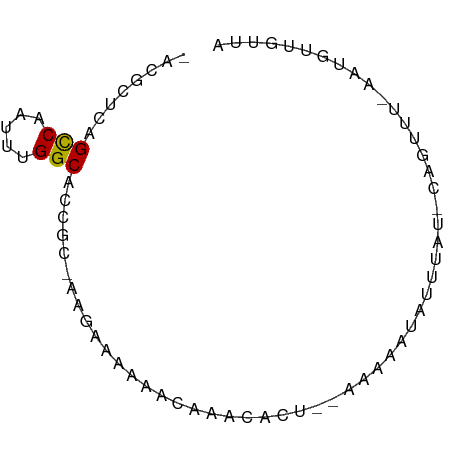
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:23:40 2011