
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,793,233 – 10,793,340 |
| Length | 107 |
| Max. P | 0.589919 |

| Location | 10,793,233 – 10,793,340 |
|---|---|
| Length | 107 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 73.37 |
| Shannon entropy | 0.57966 |
| G+C content | 0.68402 |
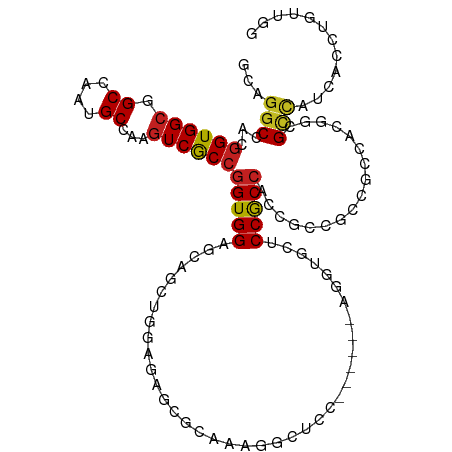
| Mean single sequence MFE | -46.20 |
| Consensus MFE | -18.62 |
| Energy contribution | -18.60 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.589919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
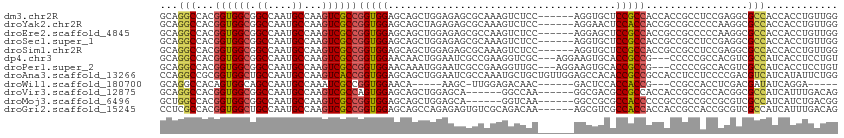
>dm3.chr2R 10793233 107 - 21146708 GCAGGCCACGGUGGCGGCCAAUGCCAAGUCGCCGGUGGAGCAGCUGGAGAGCGCAAAGUCUCC------AGGUGCUCCGCCACCACCGCCUCCGAGGCGCCACCACCUGUUGG (((((....(((((((((....))).....((((((((((((.(((((((.(.....))))))------)).))))))))).....((....)).))))))))).)))))... ( -59.70, z-score = -3.92, R) >droYak2.chr2R 10722773 107 - 21139217 GCAGGCCACGGUGGCGGCCAAUGCCAAGUCGCCGGUGGAGCAGCUAGAGAGCGCAAAGUCUCC------AGGAACUCCACCACCGCCGCCCCCAAGGCGCCACCACCUGUUGG (((((....(((((((((....((......)).(((((((...((.((((.(.....))))).------))...)))))))...)))(((.....))))))))).)))))... ( -45.30, z-score = -1.54, R) >droEre2.scaffold_4845 7529740 107 + 22589142 GCAGGCCACGGUGGCGGCCAAUGCCAAGUCGCCGGUGGAGCAGCUGGAGAGCGCCAAGUCUCC------AGGAGCUCCGCCACCGCCGCCCCCAAGGCGCCACCACCUGUUGG (((((....(((((((((....((......)).((((((((..(((((((.(.....))))))------))..))))))))...)))(((.....))))))))).)))))... ( -55.80, z-score = -2.86, R) >droSec1.super_1 8306803 107 - 14215200 GCAGGCCACGGUGGCGGCCAAUGCCAAGUCGCCGGUGGAGCAGCUGGAGAGCGCAAAGUCUCC------AGGUGCUCCGCCACCGCCGCCUCCGAGGCGCCACCACCUGUUGG (((((....(((((((((....((......)).(((((((((.(((((((.(.....))))))------)).)))))))))...)))(((.....))))))))).)))))... ( -60.60, z-score = -3.83, R) >droSim1.chr2R 9553394 107 - 19596830 GCAGGCCACGGUGGCGGCCAAUGCCAAGUCGCCGGUGGAGCAGCUGGAGAGCGCAAAGUCUCC------AGGUGCUCCGCCACCGCCGCCUCCGAGGCGCCACCACCUGUUGG (((((....(((((((((....((......)).(((((((((.(((((((.(.....))))))------)).)))))))))...)))(((.....))))))))).)))))... ( -60.60, z-score = -3.83, R) >dp4.chr3 13283724 107 - 19779522 GCAGGCCACGGUGGCGGCCAAUGCCAAGUCGCCGGUGGAACAACUGGAAUCGCCGAAGGUCGC---AGGAAGUGCACCGCCG---CCCCCGCCACGUCGCCAUCACCUCCUGU (((((....(((((((((....((...(.((.(((((.(....(((..(((......)))..)---))....).))))).))---.)...))...)))))))))....))))) ( -38.40, z-score = 0.93, R) >droPer1.super_2 697706 107 + 9036312 GCAGGCCACGGUGGCGGCCAAUGCCAAGUCGCCGGUGGAACAAAUGGAAUCGCCGAAGGUUGC---AGGAAGUGCACCGCCG---CCCCCGCCACGUCGCCAUCACCUCCUGU (((((....(((((((((....((...(.((.(((((.(.....((.((((......)))).)---).....).))))).))---.)...))...)))))))))....))))) ( -38.60, z-score = 0.68, R) >droAna3.scaffold_13266 17354075 113 + 19884421 CCAGGCCGCGGUGGCUGCCAAUGCCAAGUCACCGGUGGAGCAGCUGGAAUCGCCAAAUGCUGCUGUUGGAGCCACACCGCCGCCACCUCCUCCCCGACGUCAUCAUAUUCUGG ((((((.(((((((((.((((((((........)))..(((((((((.....)))...)))))))))))))))..))))).)))((.((......)).))..........))) ( -40.40, z-score = -0.42, R) >droWil1.scaffold_180700 3657632 93 + 6630534 GCAGGCCACAGUGGCAGCCAAUGCCAAAUCGCCGGUGGAACA-----AAGC-UUGGAGACAAC------GACUCCACCACCG---CCGCCACCUCGACGAUAUCAGGA----- ((.(((.....(((((.....)))))....)))((((((...-----..(.-(((....))))------...))))))...)---).....(((.((.....))))).----- ( -29.90, z-score = -1.65, R) >droVir3.scaffold_12875 9635629 101 - 20611582 GCAGGCCACGGUGGCGGCCAAUGCCAAGUCGCCAGUGGAGCAGCUGGAGCA------GGCCAA------GGCGACGCCGCCACCACCGCCGCCACGGCGCCAUCAUUUGACAG ...(((...(((((((((...((((..(((.(((((......)))))....------)))...------))))..)))))))))...((((...)))))))............ ( -47.00, z-score = -1.95, R) >droMoj3.scaffold_6496 9405989 101 + 26866924 GCUGGCCACGGUGGCGGCCAAUGCCAAGUCGCCGGUGGAGCAGCUGGAGCA------GGUCAA------GGCCGCGCCACCCCCGCCGCCGCCGCGUCGCCAUCAUCUGACGG (.(((((.((((((((((....((......)).(((((.((.((....)).------(((...------.))))).)))))...)))))))))).)..)))).)......... ( -46.70, z-score = 0.04, R) >droGri2.scaffold_15245 2845911 107 - 18325388 CCUCGCCACGGUGGCUGCCAAUGCCAAGUCGCCGGUGGAGCAGCCAGAGAGUGUCGCAGACAA------AGCGUCGCCACCACCACCGCCACCGCGUCGCCAUCAUUUGACAG ....((...(((((((((....))..)))))))(((((..........(.(((.(((......------.))).))))..........)))))))((((........)))).. ( -31.35, z-score = 0.82, R) >consensus GCAGGCCACGGUGGCGGCCAAUGCCAAGUCGCCGGUGGAGCAGCUGGAGAGCGCAAAGGCUCC______AGGUGCUCCGCCACCGCCGCCGCCACGGCGCCAUCACCUGUUGG ...(((...((((((.((....))...))))))(((((......................................))))).................)))............ (-18.62 = -18.60 + -0.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:23:38 2011