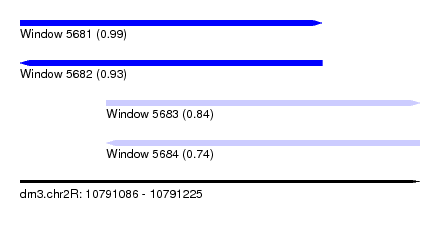
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,791,086 – 10,791,225 |
| Length | 139 |
| Max. P | 0.990252 |

| Location | 10,791,086 – 10,791,191 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 66.95 |
| Shannon entropy | 0.73861 |
| G+C content | 0.51246 |
| Mean single sequence MFE | -30.33 |
| Consensus MFE | -15.38 |
| Energy contribution | -15.55 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.990252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10791086 105 + 21146708 UUGAUAAGUACGACAGAUUGAUGGUUUUU--ACGGC-AAAUCCUAAACCCACGUUACUAGCAGAGCCGAGCAUCCGCUGACCAGCAGCGGAUAGCACGUGAAACUGCU ......((((((.(.....)..(((((..--..((.-....)).)))))..)).))))(((((..(((.((((((((((.....)))))))).)).)).)...))))) ( -28.60, z-score = -1.84, R) >droSim1.chr2R 9551297 105 + 19596830 UUGAUAAGUACGACAGAUUGGUGGCUUCU--ACGGC-UAAUCCUAAACCCACGUUACUAGCAGAGCCGAGCAUCCGCUGACCAGCAGCGGAUAGCACGUGAAACUGCU ......((((((...(((((((.(.....--.).))-))))).........)).))))(((((..(((.((((((((((.....)))))))).)).)).)...))))) ( -31.80, z-score = -1.66, R) >droSec1.super_1 8304706 105 + 14215200 UUGAUAAGUACGACAGAUUGGUGGCUUCU--ACGGC-UAAUCCUAAACCCACGUUACUAGCAGAGCCGAGCAUCCGCUGACCAGCAGCGGAUAGCACGUGAAACUGCU ......((((((...(((((((.(.....--.).))-))))).........)).))))(((((..(((.((((((((((.....)))))))).)).)).)...))))) ( -31.80, z-score = -1.66, R) >droEre2.scaffold_4845 7527578 105 - 22589142 UUGAUAAGUACGGCAGCUUGGUGGUUCUU--ACGGA-AAAUCCUAAACCCACGUUACAAGCAGAGCCGAGCAUCCGCUGACCAGCAGCGGAUAGCACGUAAAACUGCU ......((((((((.(((((((((...((--(.((.-....)))))..))))....)))))...)))).((((((((((.....)))))))).)).........)))) ( -34.80, z-score = -2.48, R) >droYak2.chr2R 10720591 107 + 21139217 UUGAUAAGUACGGCAGUUUGGUGGUGUCGGAAAGAC-AAAUCCUAAACCCACGUUACAAGCAGAGCCGAGCAUCCGCUGACCAGCAGCGGAUAGCACGUGAAACUGCU .......(((((...((((((...((((.....)))-)....))))))...)).))).(((((..(((.((((((((((.....)))))))).)).)).)...))))) ( -37.40, z-score = -2.91, R) >droAna3.scaffold_13266 17352045 107 - 19884421 -UGAUUAUUAUUGGGGUGUAUGGAGGUAUAAGUCUCAUGGAGGGCAACCCACGUUAGCAGCAAAGCCGAGCAUCCACUGACCAGCAGUGGAUAGCACGUGAAACUGCU -..........((((.(((...((((......)))).......))).))))....(((((.....(((.((((((((((.....)))))))).)).)).)...))))) ( -31.80, z-score = -0.62, R) >dp4.chr3 13281651 94 + 19779522 UUAUUGAGAAG--ACGAGAGUCAAA---------GC-CAGAA--AGACCCACGUUAGAACCAGGGCCGAGCACCCGCUGACCAGCAGCGGGUAGCACGUGAAGCUGCU ..........(--((....)))..(---------((-.....--...(((............))).((.((((((((((.....)))))))).)).))....)))... ( -28.90, z-score = -2.38, R) >droPer1.super_2 695627 94 - 9036312 UUAUUGAGAAG--GCGAGAGUCAAA---------GC-CAGAA--AGACCCACGUUAGCACCAGGGCCGAGCACCCGCUGACCAGCAGCGGGUAGCACGUGAAGCUGCU ..........(--((....)))..(---------((-.((..--.....(((((..((......))...((((((((((.....)))))))).)))))))...))))) ( -29.50, z-score = -1.33, R) >droWil1.scaffold_180700 3655578 96 - 6630534 UUAAUUUUAUACUCAAAGUGAAAGA------------CUCUAGUUUACAGACGUUAAAACUAAGGCUGAGCAUCCGCUGACCAGCAGCGGAUAACAGGUAAAGCCACU ....((((((.......))))))..------------...((((((((....))..)))))).((((..((((((((((.....)))))))).....))..))))... ( -22.30, z-score = -1.58, R) >droVir3.scaffold_12875 9633146 102 + 20611582 UUAGUCAGAAGAUUCAUCGAUGUAA-ACU---CUGC-UCUGA-AGGACGCACGUUAGCAACAGCGCUGAGCAGCCGCUGACCACCAGUGGGUAGCAGGUGAAGCUGCU ..((.(((....((((((.......-...---((((-((...-..(((....)))(((......)))))))))((((((.....))))))......)))))).))))) ( -29.90, z-score = 0.53, R) >droMoj3.scaffold_6496 9402507 97 - 26866924 UUAAUGAAAAUAUUGACAAAUUCGA---------GC-CAAGG-GCGACGCACGUUAGGAGCAGCACGGAGCAGCCGCUGACCACCAGCGGGUAGCAGGUGAAGCUGCU ((((((......((((.....))))---------((-(...)-))......)))))).((((((.....(((.((((((.....)))))).).)).......)))))) ( -27.30, z-score = -0.11, R) >droGri2.scaffold_15245 2843379 102 + 18325388 UUACUCAAU---UUUGUGGAUAUGGGACU--AUUGC-UAUUAUAGAACCCACGUUAGCAGGAGCGCUGAACAGCCGCUGACCACCAGUGGGUAGCAGGUGAAGCUGCU ........(---(((((.((..((((.((--((...-....))))..))))..)).))))))(((((.....(((((((.(((....))).)))).)))..))).)). ( -29.90, z-score = -0.51, R) >consensus UUAAUAAGUACGUCAGAGUGUUGGA_UCU__ACGGC_UAAUACUAAACCCACGUUAGAAGCAGAGCCGAGCAUCCGCUGACCAGCAGCGGAUAGCACGUGAAACUGCU .................................................((((((........)))...((((((((((.....)))))))).))..)))........ (-15.38 = -15.55 + 0.18)

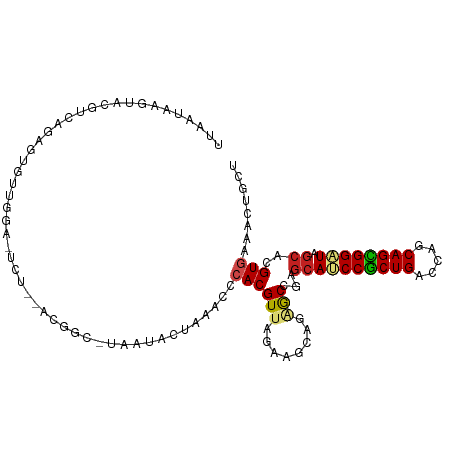
| Location | 10,791,086 – 10,791,191 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 66.95 |
| Shannon entropy | 0.73861 |
| G+C content | 0.51246 |
| Mean single sequence MFE | -30.69 |
| Consensus MFE | -15.43 |
| Energy contribution | -14.88 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.68 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.934164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10791086 105 - 21146708 AGCAGUUUCACGUGCUAUCCGCUGCUGGUCAGCGGAUGCUCGGCUCUGCUAGUAACGUGGGUUUAGGAUUU-GCCGU--AAAAACCAUCAAUCUGUCGUACUUAUCAA (((((.....((.((.((((((((.....)))))))))).))...)))))((((.((((((((..((.(((-.....--.))).))...)))))).))))))...... ( -32.00, z-score = -1.46, R) >droSim1.chr2R 9551297 105 - 19596830 AGCAGUUUCACGUGCUAUCCGCUGCUGGUCAGCGGAUGCUCGGCUCUGCUAGUAACGUGGGUUUAGGAUUA-GCCGU--AGAAGCCACCAAUCUGUCGUACUUAUCAA (((((.....((.((.((((((((.....)))))))))).))...)))))......((((.(((.((....-.))..--.))).)))).................... ( -33.10, z-score = -0.61, R) >droSec1.super_1 8304706 105 - 14215200 AGCAGUUUCACGUGCUAUCCGCUGCUGGUCAGCGGAUGCUCGGCUCUGCUAGUAACGUGGGUUUAGGAUUA-GCCGU--AGAAGCCACCAAUCUGUCGUACUUAUCAA (((((.....((.((.((((((((.....)))))))))).))...)))))......((((.(((.((....-.))..--.))).)))).................... ( -33.10, z-score = -0.61, R) >droEre2.scaffold_4845 7527578 105 + 22589142 AGCAGUUUUACGUGCUAUCCGCUGCUGGUCAGCGGAUGCUCGGCUCUGCUUGUAACGUGGGUUUAGGAUUU-UCCGU--AAGAACCACCAAGCUGCCGUACUUAUCAA ...........(((((((((((((.....)))))))))...(((...(((((....((((.((((((....-.)).)--)))..))))))))).)))))))....... ( -35.20, z-score = -1.58, R) >droYak2.chr2R 10720591 107 - 21139217 AGCAGUUUCACGUGCUAUCCGCUGCUGGUCAGCGGAUGCUCGGCUCUGCUUGUAACGUGGGUUUAGGAUUU-GUCUUUCCGACACCACCAAACUGCCGUACUUAUCAA (((((.....((.((.((((((((.....)))))))))).))...))))).(((.((..(((((.((...(-(((.....))))...)))))))..)))))....... ( -36.10, z-score = -2.16, R) >droAna3.scaffold_13266 17352045 107 + 19884421 AGCAGUUUCACGUGCUAUCCACUGCUGGUCAGUGGAUGCUCGGCUUUGCUGCUAACGUGGGUUGCCCUCCAUGAGACUUAUACCUCCAUACACCCCAAUAAUAAUCA- ((((((....((.((.((((((((.....)))))))))).)).....))))))..((((((......))))))..................................- ( -31.40, z-score = -1.72, R) >dp4.chr3 13281651 94 - 19779522 AGCAGCUUCACGUGCUACCCGCUGCUGGUCAGCGGGUGCUCGGCCCUGGUUCUAACGUGGGUCU--UUCUG-GC---------UUUGACUCUCGU--CUUCUCAAUAA ((.((((...((.((.((((((((.....)))))))))).)).....)))))).(((.(((((.--.....-..---------...))))).)))--........... ( -30.40, z-score = -1.18, R) >droPer1.super_2 695627 94 + 9036312 AGCAGCUUCACGUGCUACCCGCUGCUGGUCAGCGGGUGCUCGGCCCUGGUGCUAACGUGGGUCU--UUCUG-GC---------UUUGACUCUCGC--CUUCUCAAUAA ((((.(....((.((.((((((((.....)))))))))).)).....).))))..((.(((((.--.....-..---------...))))).)).--........... ( -33.00, z-score = -1.05, R) >droWil1.scaffold_180700 3655578 96 + 6630534 AGUGGCUUUACCUGUUAUCCGCUGCUGGUCAGCGGAUGCUCAGCCUUAGUUUUAACGUCUGUAAACUAGAG------------UCUUUCACUUUGAGUAUAAAAUUAA ...((((......((.((((((((.....))))))))))..)))).((((((((((((......))(((((------------(.....)))))).)).)))))))). ( -22.10, z-score = -0.56, R) >droVir3.scaffold_12875 9633146 102 - 20611582 AGCAGCUUCACCUGCUACCCACUGGUGGUCAGCGGCUGCUCAGCGCUGUUGCUAACGUGCGUCCU-UCAGA-GCAG---AGU-UUACAUCGAUGAAUCUUCUGACUAA (((((((....((((((((....))))).))).)))))))..(((((((.....))).))))...-(((((-..((---(.(-(((......)))))))))))).... ( -28.30, z-score = 0.76, R) >droMoj3.scaffold_6496 9402507 97 + 26866924 AGCAGCUUCACCUGCUACCCGCUGGUGGUCAGCGGCUGCUCCGUGCUGCUCCUAACGUGCGUCGC-CCUUG-GC---------UCGAAUUUGUCAAUAUUUUCAUUAA (((((((....((((((((....))))).))).)))))))..(((.(((..(....).))).)))-..(((-((---------........)))))............ ( -26.80, z-score = -0.27, R) >droGri2.scaffold_15245 2843379 102 - 18325388 AGCAGCUUCACCUGCUACCCACUGGUGGUCAGCGGCUGUUCAGCGCUCCUGCUAACGUGGGUUCUAUAAUA-GCAAU--AGUCCCAUAUCCACAAA---AUUGAGUAA .((((((....((((((((....))))).))).))))))((((.((....))....(((((..((((....-...))--)).))))).........---.)))).... ( -26.80, z-score = -0.30, R) >consensus AGCAGCUUCACGUGCUAUCCGCUGCUGGUCAGCGGAUGCUCGGCUCUGCUGCUAACGUGGGUUUAGGAUUA_GCCGU__AGA_UCCAACAAUCUGACGUACUUAUCAA ((((((.......)))((((((((.....))))))))))).(((((............)))))............................................. (-15.43 = -14.88 + -0.56)
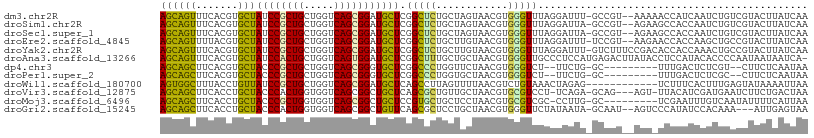
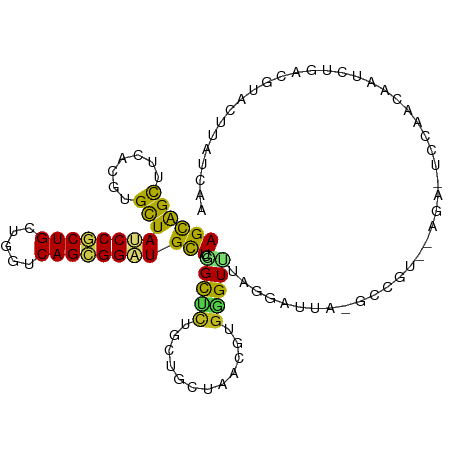
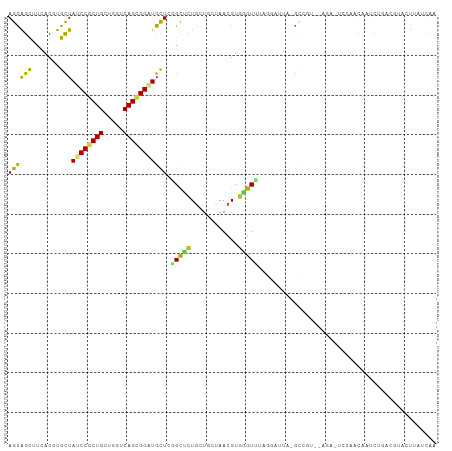
| Location | 10,791,116 – 10,791,225 |
|---|---|
| Length | 109 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.45 |
| Shannon entropy | 0.45369 |
| G+C content | 0.53919 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -21.11 |
| Energy contribution | -21.16 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.843149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10791116 109 + 21146708 --CGGCAAAUCCUAAACCCACGUUACUAGCAGAGCCGAGCAUCCGCUGACCAGCAGCGGAUAGCACGUGAAACUGCUCAGGCUGGAAAACAUUGGCAGCACCAUGCGAAAA----- --((((........(((....))).(((((((..(((.((((((((((.....)))))))).)).)).)...))))).)))))).......((.(((......))).))..----- ( -33.50, z-score = -1.41, R) >droSim1.chr2R 9551327 109 + 19596830 --CGGCUAAUCCUAAACCCACGUUACUAGCAGAGCCGAGCAUCCGCUGACCAGCAGCGGAUAGCACGUGAAACUGCUCAGGCUGGAAAACAUUGGCAGCACCAUGCGAAAA----- --..((((((.......(((.(((...(((((..(((.((((((((((.....)))))))).)).)).)...)))))..)))))).....)))))).((.....)).....----- ( -34.50, z-score = -1.62, R) >droSec1.super_1 8304736 109 + 14215200 --CGGCUAAUCCUAAACCCACGUUACUAGCAGAGCCGAGCAUCCGCUGACCAGCAGCGGAUAGCACGUGAAACUGCUCAGGCUGGAAAACAUUGGCAGCACCAUGCGAAAA----- --..((((((.......(((.(((...(((((..(((.((((((((((.....)))))))).)).)).)...)))))..)))))).....)))))).((.....)).....----- ( -34.50, z-score = -1.62, R) >droEre2.scaffold_4845 7527608 109 - 22589142 --CGGAAAAUCCUAAACCCACGUUACAAGCAGAGCCGAGCAUCCGCUGACCAGCAGCGGAUAGCACGUAAAACUGCUCAGGCUGGAAAACAUUGGCAGCACCAUGCGAAAA----- --.((.....)).....(((.(((...(((((...((.((((((((((.....)))))))).)).)).....)))))..))))))......((.(((......))).))..----- ( -32.00, z-score = -1.77, R) >droYak2.chr2R 10720622 110 + 21139217 -AAGACAAAUCCUAAACCCACGUUACAAGCAGAGCCGAGCAUCCGCUGACCAGCAGCGGAUAGCACGUGAAACUGCUCAGGCUGGAGAACAUUGGCAGCACCAUGCGAAAA----- -................(((.(((...(((((..(((.((((((((((.....)))))))).)).)).)...)))))..))))))......((.(((......))).))..----- ( -32.30, z-score = -1.82, R) >droAna3.scaffold_13266 17352081 105 - 19884421 --UGGAGGGC----AACCCACGUUAGCAGCAAAGCCGAGCAUCCACUGACCAGCAGUGGAUAGCACGUGAAACUGCUCAGGCUGGAAAACAUUGGCACCACCAUGCGGAAA----- --(((.((((----(...(((((..((......))...((((((((((.....)))))))).)))))))....)))))..((..(......)..)).)))((....))...----- ( -33.10, z-score = -0.76, R) >dp4.chr3 13281676 103 + 19779522 --------CAGAAAGACCCACGUUAGAACCAGGGCCGAGCACCCGCUGACCAGCAGCGGGUAGCACGUGAAGCUGCUCAGGCUGGAGAACAUUGGCAAAACCAUUCGGAAA----- --------.........((..(((....(((..(((((((((((((((.....))))))))(((.......))))))).))))))..)))..(((.....)))...))...----- ( -34.70, z-score = -1.72, R) >droPer1.super_2 695652 103 - 9036312 --------CAGAAAGACCCACGUUAGCACCAGGGCCGAGCACCCGCUGACCAGCAGCGGGUAGCACGUGAAGCUGCUCAGGCUGGAGAACAUUGGCAAAACCAUUCGGAAA----- --------.........((..(((....(((..(((((((((((((((.....))))))))(((.......))))))).))))))..)))..(((.....)))...))...----- ( -34.70, z-score = -1.32, R) >droWil1.scaffold_180700 3655597 111 - 6630534 GAAAGACUCUAGUUUACAGACGUUAAAACUAAGGCUGAGCAUCCGCUGACCAGCAGCGGAUAACAGGUAAAGCCACUCAGACUGGAAAACAUUGGCACAACCAUUCAGAAA----- ......(..((((((((....))..))))))..)(((((.((((((((.....))))))))....((.....)).))))).(((((......(((.....))))))))...----- ( -28.60, z-score = -2.32, R) >droVir3.scaffold_12875 9633177 104 + 20611582 ------CUCUGAAGGACGCACGUUAGCAACAGCGCUGAGCAGCCGCUGACCACCAGUGGGUAGCAGGUGAAGCUGCUCAGGCCGGAAAACAUCGAGAACACCAUGGAAAA------ ------.((((......((......))....((.(((((((((((((..(.(((....))).)..))))..)))))))))))))))........................------ ( -30.80, z-score = 0.15, R) >droGri2.scaffold_15245 2843409 111 + 18325388 -----CUAUUAUAGAACCCACGUUAGCAGGAGCGCUGAACAGCCGCUGACCACCAGUGGGUAGCAGGUGAAGCUGCUCAGGCCGGAAAACAUCGAGAACACCAUCGAGAACACAAA -----................(((.((..(((((((.....(((((((.(((....))).)))).)))..))).))))..)).........((((........)))).)))..... ( -29.70, z-score = -0.82, R) >consensus ___GGCAAAUCCUAAACCCACGUUACAAGCAGAGCCGAGCAUCCGCUGACCAGCAGCGGAUAGCACGUGAAACUGCUCAGGCUGGAAAACAUUGGCAGCACCAUGCGAAAA_____ .................................(((((((((((((((.....)))))))).))..((....(.((....)).)....)).))))).................... (-21.11 = -21.16 + 0.05)

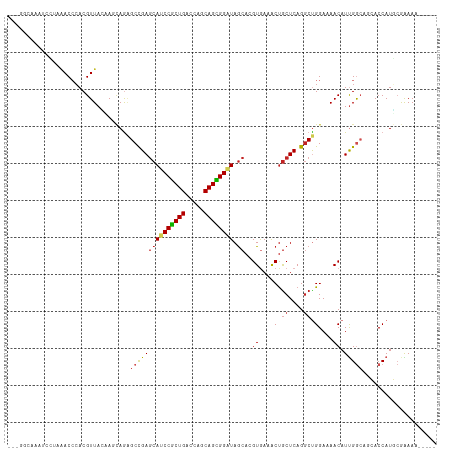
| Location | 10,791,116 – 10,791,225 |
|---|---|
| Length | 109 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 78.45 |
| Shannon entropy | 0.45369 |
| G+C content | 0.53919 |
| Mean single sequence MFE | -35.78 |
| Consensus MFE | -26.79 |
| Energy contribution | -26.12 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.39 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.743177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10791116 109 - 21146708 -----UUUUCGCAUGGUGCUGCCAAUGUUUUCCAGCCUGAGCAGUUUCACGUGCUAUCCGCUGCUGGUCAGCGGAUGCUCGGCUCUGCUAGUAACGUGGGUUUAGGAUUUGCCG-- -----.....(((.(((.(((((.(((((..(.(((..((((.......((.((.((((((((.....)))))))))).)))))).))).).))))).)))..)).))))))..-- ( -35.71, z-score = -0.12, R) >droSim1.chr2R 9551327 109 - 19596830 -----UUUUCGCAUGGUGCUGCCAAUGUUUUCCAGCCUGAGCAGUUUCACGUGCUAUCCGCUGCUGGUCAGCGGAUGCUCGGCUCUGCUAGUAACGUGGGUUUAGGAUUAGCCG-- -----.....((.((((.(((((.(((((..(.(((..((((.......((.((.((((((((.....)))))))))).)))))).))).).))))).)))..)).))))))..-- ( -36.11, z-score = -0.18, R) >droSec1.super_1 8304736 109 - 14215200 -----UUUUCGCAUGGUGCUGCCAAUGUUUUCCAGCCUGAGCAGUUUCACGUGCUAUCCGCUGCUGGUCAGCGGAUGCUCGGCUCUGCUAGUAACGUGGGUUUAGGAUUAGCCG-- -----.....((.((((.(((((.(((((..(.(((..((((.......((.((.((((((((.....)))))))))).)))))).))).).))))).)))..)).))))))..-- ( -36.11, z-score = -0.18, R) >droEre2.scaffold_4845 7527608 109 + 22589142 -----UUUUCGCAUGGUGCUGCCAAUGUUUUCCAGCCUGAGCAGUUUUACGUGCUAUCCGCUGCUGGUCAGCGGAUGCUCGGCUCUGCUUGUAACGUGGGUUUAGGAUUUUCCG-- -----.....((((((.....)).))))..((((((((((((((.....((.((.((((((((.....)))))))))).))...)))))).......)))))..))).......-- ( -35.01, z-score = -0.68, R) >droYak2.chr2R 10720622 110 - 21139217 -----UUUUCGCAUGGUGCUGCCAAUGUUCUCCAGCCUGAGCAGUUUCACGUGCUAUCCGCUGCUGGUCAGCGGAUGCUCGGCUCUGCUUGUAACGUGGGUUUAGGAUUUGUCUU- -----.....((((((.....)).))))..((((((((((((((.....((.((.((((((((.....)))))))))).))...)))))).......)))))..)))........- ( -34.91, z-score = -0.22, R) >droAna3.scaffold_13266 17352081 105 + 19884421 -----UUUCCGCAUGGUGGUGCCAAUGUUUUCCAGCCUGAGCAGUUUCACGUGCUAUCCACUGCUGGUCAGUGGAUGCUCGGCUUUGCUGCUAACGUGGGUU----GCCCUCCA-- -----........(((.((.(((.(((((...(((((((((((((.......)))((((((((.....))))))))))))))....))))..))))).))).----..)).)))-- ( -37.50, z-score = -1.07, R) >dp4.chr3 13281676 103 - 19779522 -----UUUCCGAAUGGUUUUGCCAAUGUUCUCCAGCCUGAGCAGCUUCACGUGCUACCCGCUGCUGGUCAGCGGGUGCUCGGCCCUGGUUCUAACGUGGGUCUUUCUG-------- -----..(((...(((.....)))(((((..((((.(((((((((.......)))((((((((.....))))))))))))))..))))....))))))))........-------- ( -37.90, z-score = -1.95, R) >droPer1.super_2 695652 103 + 9036312 -----UUUCCGAAUGGUUUUGCCAAUGUUCUCCAGCCUGAGCAGCUUCACGUGCUACCCGCUGCUGGUCAGCGGGUGCUCGGCCCUGGUGCUAACGUGGGUCUUUCUG-------- -----..(((...(((.....)))((((((.((((.(((((((((.......)))((((((((.....))))))))))))))..)))).)..))))))))........-------- ( -37.90, z-score = -1.39, R) >droWil1.scaffold_180700 3655597 111 + 6630534 -----UUUCUGAAUGGUUGUGCCAAUGUUUUCCAGUCUGAGUGGCUUUACCUGUUAUCCGCUGCUGGUCAGCGGAUGCUCAGCCUUAGUUUUAACGUCUGUAAACUAGAGUCUUUC -----.....((.(((((.(((..(((((....((.((((((((.....))....((((((((.....)))))))))))))).)).......)))))..))))))))...)).... ( -28.80, z-score = -1.07, R) >droVir3.scaffold_12875 9633177 104 - 20611582 ------UUUUCCAUGGUGUUCUCGAUGUUUUCCGGCCUGAGCAGCUUCACCUGCUACCCACUGGUGGUCAGCGGCUGCUCAGCGCUGUUGCUAACGUGCGUCCUUCAGAG------ ------.................(((((....((((((((((((((....((((((((....))))).))).)))))))))).))))..((....)))))))........------ ( -37.40, z-score = -1.71, R) >droGri2.scaffold_15245 2843409 111 - 18325388 UUUGUGUUCUCGAUGGUGUUCUCGAUGUUUUCCGGCCUGAGCAGCUUCACCUGCUACCCACUGGUGGUCAGCGGCUGUUCAGCGCUCCUGCUAACGUGGGUUCUAUAAUAG----- .........((((........))))........(((((((((((((....((((((((....))))).))).)))))))))).)))((..(....)..))...........----- ( -36.20, z-score = -1.38, R) >consensus _____UUUUCGCAUGGUGCUGCCAAUGUUUUCCAGCCUGAGCAGUUUCACGUGCUAUCCGCUGCUGGUCAGCGGAUGCUCGGCUCUGCUGCUAACGUGGGUUUAGGAUUAGCC___ ....................(((.(((((...(((.(((((((((.......)))((((((((.....))))))))))))))..))).....))))).)))............... (-26.79 = -26.12 + -0.67)
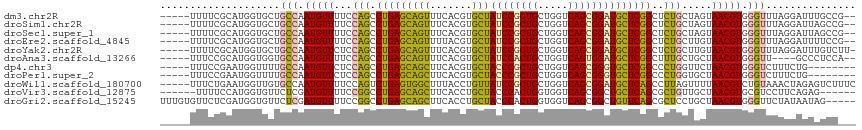

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:23:37 2011