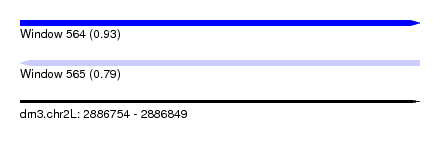
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,886,754 – 2,886,849 |
| Length | 95 |
| Max. P | 0.934805 |

| Location | 2,886,754 – 2,886,849 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 77.26 |
| Shannon entropy | 0.43008 |
| G+C content | 0.43786 |
| Mean single sequence MFE | -24.86 |
| Consensus MFE | -16.68 |
| Energy contribution | -16.60 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934805 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
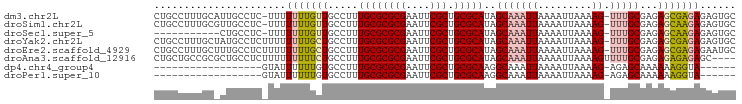
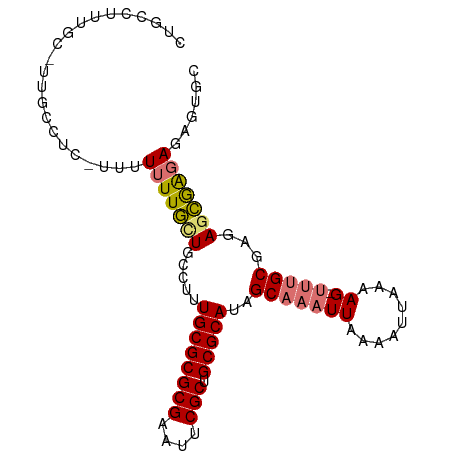
>dm3.chr2L 2886754 95 + 23011544 GCACUCUCUCGCUCUCGCAAA-CUUUUAAUUUUAAUUUGCUAUGCGCAGCGAAUUCGCGCGCAAAGGCAACAAAAAAA-GAGGCAAUGCAAAGGCAG ((.((((...((....))...-..............(((((.(((((.(((....))))))))..))))).......)-)))))..(((....))). ( -26.60, z-score = -1.43, R) >droSim1.chr2L 2847444 95 + 22036055 GCACUCUCUUGCUCUCGCAAA-CUUUUAAUUUUAAUUUGCUAUGCGCAGCGAAUUCGCGCGCAAAGGCAACAAAAAAA-GAGGCAACGCAAAGGCAG ((.((((.((((....)))).-..............(((((.(((((.(((....))))))))..))))).......)-)))))...((....)).. ( -27.30, z-score = -1.64, R) >droSec1.super_5 1045559 84 + 5866729 GCACUCUCUUGCUCUCGCAAA-CUUUUAAUUUUAAUUUGCUAUGCGCAGCGAAUUCGCGCGCAAAGGCAACAAAAAAA-GAGGCAG----------- ((.((((.((((....)))).-..............(((((.(((((.(((....))))))))..))))).......)-)))))..----------- ( -26.10, z-score = -2.56, R) >droYak2.chr2L 2881826 96 + 22324452 GCACUCUCUCGCUCUCGCAAA-CUUUUAAUUUUAAUUUGCUAUGCGCAGCGAAUUCGCGCGCAAAGGCAGCAAAAAAAAGAGGCAUAGCAAAGGCAG ((........((....))...-.............((((((((((((.(((....)))))((....))..............)))))))))).)).. ( -25.90, z-score = -0.58, R) >droEre2.scaffold_4929 2933984 96 + 26641161 GCAUUCUCUCGCUCUCGCAAA-CUUUUAAUUUUAAUUUGCUAUGCGCAGCGAAUUCGCGCGCAAAGGCAGCAAAAAAAAGAGGCAAAGCAAAGGCAG ((.((.((((((....))...-.............((((((.(((((.(((....)))))))).....)))))).....)))).)).))........ ( -24.50, z-score = -0.06, R) >droAna3.scaffold_12916 748996 93 + 16180835 ----GCUCUCUCUCUCGCAAAACUUUUAAUUUUAAUUUGCUAUGCGCAGCGAAUUCGCGCGCAAAGGCAGAAAAAAAAAGAGGCAGCGCGGCAGCAG ----(((.(((((.....((((.......))))..((((((.(((((.(((....))))))))..)))))).......))))).)))((....)).. ( -26.70, z-score = -0.26, R) >dp4.chr4_group4 2284901 72 - 6586962 ------UACCUUUUUUGCUCU-CUUUUAAUUUUAAUUUGCCUUGCGCAGCGAAUUCGCGCGCAAAGGCACAAAAAAUAC------------------ ------....(((((((....-...............((((((((((.(((....))))))).)))))))))))))...------------------ ( -20.90, z-score = -3.52, R) >droPer1.super_10 90564 72 - 3432795 ------UACCUUUUUUGCUCU-CUUUUAAUUUUAAUUUGCCUUGCGCAGCGAAUUCGCGCGCAAAGGCACAAAAAAUAC------------------ ------....(((((((....-...............((((((((((.(((....))))))).)))))))))))))...------------------ ( -20.90, z-score = -3.52, R) >consensus GCACUCUCUCGCUCUCGCAAA_CUUUUAAUUUUAAUUUGCUAUGCGCAGCGAAUUCGCGCGCAAAGGCAACAAAAAAA_GAGGCAA_GCAAAGGCAG ....................................(((((.(((((.(((....))))))))..)))))........................... (-16.68 = -16.60 + -0.08)

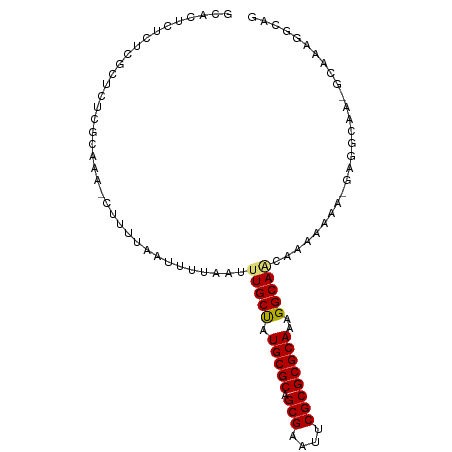
| Location | 2,886,754 – 2,886,849 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 77.26 |
| Shannon entropy | 0.43008 |
| G+C content | 0.43786 |
| Mean single sequence MFE | -27.44 |
| Consensus MFE | -16.92 |
| Energy contribution | -17.44 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.787683 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 2886754 95 - 23011544 CUGCCUUUGCAUUGCCUC-UUUUUUUGUUGCCUUUGCGCGCGAAUUCGCUGCGCAUAGCAAAUUAAAAUUAAAAG-UUUGCGAGAGCGAGAGAGUGC .(((....)))..(((((-((((((..((((...((((((((....))).)))))..))))...)))).......-.((((....))))))))).)) ( -29.60, z-score = -1.26, R) >droSim1.chr2L 2847444 95 - 22036055 CUGCCUUUGCGUUGCCUC-UUUUUUUGUUGCCUUUGCGCGCGAAUUCGCUGCGCAUAGCAAAUUAAAAUUAAAAG-UUUGCGAGAGCAAGAGAGUGC ..((....))...(((((-((((((..((((...((((((((....))).)))))..))))...)))).......-.((((....))))))))).)) ( -29.30, z-score = -1.11, R) >droSec1.super_5 1045559 84 - 5866729 -----------CUGCCUC-UUUUUUUGUUGCCUUUGCGCGCGAAUUCGCUGCGCAUAGCAAAUUAAAAUUAAAAG-UUUGCGAGAGCAAGAGAGUGC -----------..(((((-((((((..((((...((((((((....))).)))))..))))...)))).......-.((((....))))))))).)) ( -29.00, z-score = -2.34, R) >droYak2.chr2L 2881826 96 - 22324452 CUGCCUUUGCUAUGCCUCUUUUUUUUGCUGCCUUUGCGCGCGAAUUCGCUGCGCAUAGCAAAUUAAAAUUAAAAG-UUUGCGAGAGCGAGAGAGUGC .............(((((((((.(((((((....((((((((....))).)))))))))))).............-...((....))))))))).)) ( -30.00, z-score = -1.19, R) >droEre2.scaffold_4929 2933984 96 - 26641161 CUGCCUUUGCUUUGCCUCUUUUUUUUGCUGCCUUUGCGCGCGAAUUCGCUGCGCAUAGCAAAUUAAAAUUAAAAG-UUUGCGAGAGCGAGAGAAUGC .....((((((((((..(((((.(((((((....((((((((....))).))))))))))))........)))))-...)).))))))))....... ( -28.60, z-score = -0.90, R) >droAna3.scaffold_12916 748996 93 - 16180835 CUGCUGCCGCGCUGCCUCUUUUUUUUUCUGCCUUUGCGCGCGAAUUCGCUGCGCAUAGCAAAUUAAAAUUAAAAGUUUUGCGAGAGAGAGAGC---- ..((.((...)).))(((((((((((..(((...((((((((....))).)))))..)))...((((((.....)))))).))))))))))).---- ( -27.20, z-score = -0.03, R) >dp4.chr4_group4 2284901 72 + 6586962 ------------------GUAUUUUUUGUGCCUUUGCGCGCGAAUUCGCUGCGCAAGGCAAAUUAAAAUUAAAAG-AGAGCAAAAAAGGUA------ ------------------...(((((((((((((.(((((((....))).)))))))))...(((....)))...-...))))))))....------ ( -22.90, z-score = -2.56, R) >droPer1.super_10 90564 72 + 3432795 ------------------GUAUUUUUUGUGCCUUUGCGCGCGAAUUCGCUGCGCAAGGCAAAUUAAAAUUAAAAG-AGAGCAAAAAAGGUA------ ------------------...(((((((((((((.(((((((....))).)))))))))...(((....)))...-...))))))))....------ ( -22.90, z-score = -2.56, R) >consensus CUGCCUUUGC_UUGCCUC_UUUUUUUGCUGCCUUUGCGCGCGAAUUCGCUGCGCAUAGCAAAUUAAAAUUAAAAG_UUUGCGAGAGCGAGAGAGUGC .....................((((((((.....((((((((....))).)))))..(((((..............)))))...))))))))..... (-16.92 = -17.44 + 0.52)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:12:30 2011