| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,746,706 – 10,746,802 |
| Length | 96 |
| Max. P | 0.886497 |

| Location | 10,746,706 – 10,746,802 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 61.01 |
| Shannon entropy | 0.87171 |
| G+C content | 0.40594 |
| Mean single sequence MFE | -20.55 |
| Consensus MFE | -11.08 |
| Energy contribution | -10.60 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.92 |
| Mean z-score | -0.21 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.886497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
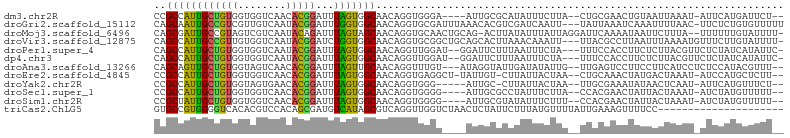
>dm3.chr2R 10746706 96 - 21146708 CCGCCAUUGCUGUGGUGGUCAACACGGAUUUAGUGGCAACAGGUGGGA----AUUGCGCAUAUUUCUUA--CUGCGAACUGUAAUUAAAU-AUUCAUGAUUCU-- (((((....(((((........)))))......((....)))))))((----(((..(.((((((.(((--(........))))..))))-)).)..))))).-- ( -24.30, z-score = -0.72, R) >droGri2.scaffold_15112 3482948 101 + 5172618 CAGCAAUUGCCGUCGUUGUCAAUACGGAUUUGGUGGCAACAGGUGCGAUUUAAACACGUCGAUCAAUU---UAUUAAAUCAAAUUUUAAC-UUCUCUGUGUUUUU ..(((.(((((..((..(((......))).))..)))))....))).....(((((((..((......---.(((......)))......-.))..))))))).. ( -18.74, z-score = -0.08, R) >droMoj3.scaffold_6496 13409832 101 - 26866924 CAGCGAUUGCCGUAGUCGUCAAUACAGAUUUAGUAGCAACAGGUGCAACUGCAG-ACUUAUAUUUAUUAGGAUUCAAAAUAAUUCUUUA--UUUUUUGUAUUUU- ..(((((((...))))))).((((((((..((((.(((.....))).))))...-.............((((((......))))))...--...))))))))..- ( -15.00, z-score = 0.19, R) >droVir3.scaffold_12875 13790206 101 + 20611582 CAGCCAUUGCCGUUGUGGUCAAUACGGAUCUGGUGGCAACAGGUGCGGCUGCAGCACUUAAACAAAUU---UUACGCCUUAAUUUAAAAUGUUUCUUGUAUUUU- ..((..((((((..(.((((......)))))..)))))).((((((.......)))))).........---....))...........................- ( -20.30, z-score = 1.13, R) >droPer1.super_4 3244123 99 + 7162766 CAGCCAUUGCUGUGGUGGUCAAUACGGAUUUAGUGGCAACAGGUUGGAU--GGAUUCUUUAAUUUCUA---UUUCCACCUUCUCUUACGUUCUCUAUCAUAUUC- ((((....))))((((((.....(((.......((....))(((.((((--(((..........))))---..))))))........)))...)))))).....- ( -20.10, z-score = -0.50, R) >dp4.chr3 15132426 99 - 19779522 CAGCCAUUGCUGUGGUGGUCAAUACGGAUUUAGUGGCAACAGGUUGGAU--GGAUUCUUUAAUUUCUA---UUUCCACCUUCUCUUACGUUCUCUAUCAUAUUC- ((((....))))((((((.....(((.......((....))(((.((((--(((..........))))---..))))))........)))...)))))).....- ( -20.10, z-score = -0.50, R) >droAna3.scaffold_13266 5347378 98 - 19884421 CAGCAGUUGCUGUGGUAGUCAACACGGAUUUAGUUGCAACAGGUUUGU---AUAGGUAUUGAUAUAUUG--UUGAGUCCUUCCUUCAUCCUCUCCAUACGUUU-- ((((....)))).((.(.(((((((((((((.((....))))))))))---...(((((....))))))--)))).)))........................-- ( -16.20, z-score = 1.25, R) >droEre2.scaffold_4845 7493429 98 + 22589142 CCGCCAUUGCUGUGGUGGUCAACACGGAUUUAGUGGCAACAGGUGAGGCU-UAUUGU-CUUAUUACUAA--CUGCAAACUAUGACUAAAU-AUCCAUGCUCUU-- (((((((....))))))).......((((((((((....).((((((((.-....))-)))))).....--............)))))..-))))........-- ( -25.50, z-score = -1.08, R) >droYak2.chr2R 10684371 94 - 21139217 CCGCCAUUGCUGUGGUAGUGAACACGGAUUUAGUGGCAACAGGUGGG-----AUUGC-CUUAUUACUAA--UUGCGAAAUAUAACUCAAU-AUUCAUGUUUCU-- ..((((((((((((........)))))...)))))))...((((...-----...))-)).........--....(((((((........-....))))))).-- ( -20.60, z-score = -0.05, R) >droSec1.super_1 8269332 96 - 14215200 CCGCCAUUGCUGUGGUGGUCAACACGGAUUUAGUGGCAACAGGUGGGG----AUUGCGCCUAUUUCUUA--CCACGAACUAUUACUAAAU-AUCUAUGUUUUU-- (((((((....)))))))..((((..(((((((((....).(((((((----(..........))))))--))..........)))))..-)))..))))...-- ( -27.20, z-score = -1.68, R) >droSim1.chr2R 9516417 96 - 19596830 CCGCUAUUGCUGUGGUGGUCAACACGGAUUUAGUGGCAACAGGUGGGG----AUUGCGUAUAUUUCUUU--CCACGAACUAUUACUAAAU-AUCUAUGUUUUU-- (((((((....)))))))..((((..(((((((((....)..((((..----(.............)..--))))........)))))..-)))..))))...-- ( -22.82, z-score = -0.94, R) >triCas2.ChLG5 11489380 84 + 18847211 GUGCCGUGGGGUCACACGUCCACAGCGAUGUCAUAGCGUCAGGUUGGUCUAACUCUAUUCUUUAUGUUUUAUUGAAAGUUUUCC--------------------- ..((((((((........)))))...(((((....))))).))).((...((((...(((..((.....))..)))))))..))--------------------- ( -15.70, z-score = 0.49, R) >consensus CAGCCAUUGCUGUGGUGGUCAACACGGAUUUAGUGGCAACAGGUGGGA____AUUGCUUUUAUUUCUUA__UUGCGAACUAUUAUUAAAU_AUCUAUGUUUUU__ ..((((((((((((........)))))...))))))).................................................................... (-11.08 = -10.60 + -0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:23:31 2011