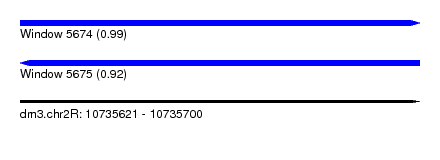
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,735,621 – 10,735,700 |
| Length | 79 |
| Max. P | 0.992093 |

| Location | 10,735,621 – 10,735,700 |
|---|---|
| Length | 79 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 73.24 |
| Shannon entropy | 0.49545 |
| G+C content | 0.44425 |
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -11.94 |
| Energy contribution | -13.67 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.992093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
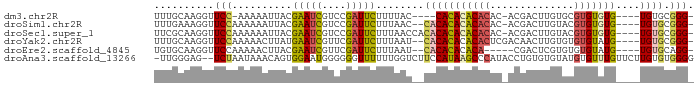
>dm3.chr2R 10735621 79 + 21146708 -CCCGCACA----CACACACGCACAAGUCGU-GUGUGUGUGUG----GUAAAAGAAUCGGACGAUUCGUAAUUUUU-GGAACCUUGCAAA -.(((((((----(((((((((....).)))-)))))))))))----).....(((((....)))))((((.....-......))))... ( -32.70, z-score = -4.21, R) >droSim1.chr2R 9505199 82 + 19596830 -CCCGCACA----CACACACGUACAAGUCGU-GUGUGUGUGUGUG--GUUAAAGAAUCGGACGAUUCGUAAUUUUUUGGAACCUUUCAAA -((((((((----((((((((.......)))-))))))))))).)--).....(((((....))))).......((((((....)))))) ( -33.00, z-score = -4.34, R) >droSec1.super_1 8258092 84 + 14215200 -CCCGCACA----CACACACGUACAAGUCGU-GUGUGUGUGUGUGUGGUUAAAGAAUCGGACGAUUCGUAAUUUUUUGGAACCUUGCGAA -.(((((((----(((((((((((.....))-)))))))))))))))).....(((((....)))))....................... ( -33.90, z-score = -3.74, R) >droYak2.chr2R 10673119 83 + 21139217 -CCCGCACA----CAUACACACACAAGUUGUCGAGUGUGUGUGUG--AUUAAAGAAUCGAACGAUUCAUAAGUUUUUGGAACCUUGCAAA -...(((..----(((((((((((..(....)..)))))))))))--......(((((....))))).................)))... ( -25.00, z-score = -2.82, R) >droEre2.scaffold_4845 7482137 78 - 22589142 -CCUGCACA----CAUACACACACGAGUCG-----UGUGUGUGUG--AUUAAAGAAUCGAACGAUUCGUAAGUUUUUGGAACCUUGCACA -..((((..----((((((((((((...))-----))))))))))--......(((((....))))).................)))).. ( -26.30, z-score = -2.92, R) >droAna3.scaffold_13266 8986350 87 + 19884421 CCCCACACAAGAACAAACACAUACACACAGGUAUGGGCUUAUGGAAGACCAAAAAACCCCCCAUUCCACUGUUUAUUAGA--CUCCCAA- ..........(((((....(((((......)))))((...((((................)))).))..)))))......--.......- ( -10.59, z-score = 0.66, R) >consensus _CCCGCACA____CACACACACACAAGUCGU_GUGUGUGUGUGUG__AUUAAAGAAUCGGACGAUUCGUAAUUUUUUGGAACCUUGCAAA ..((.........(((((((((((........)))))))))))..........(((((....)))))..........))........... (-11.94 = -13.67 + 1.72)

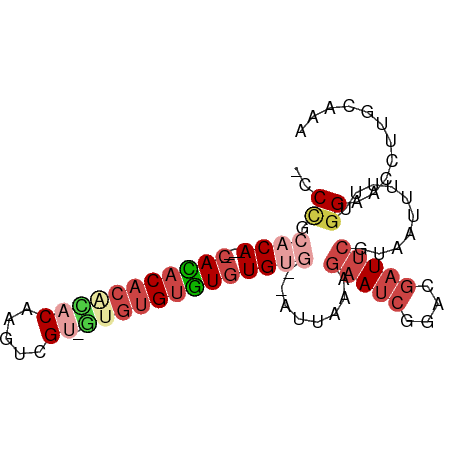
| Location | 10,735,621 – 10,735,700 |
|---|---|
| Length | 79 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 73.24 |
| Shannon entropy | 0.49545 |
| G+C content | 0.44425 |
| Mean single sequence MFE | -25.62 |
| Consensus MFE | -9.97 |
| Energy contribution | -9.89 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.59 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10735621 79 - 21146708 UUUGCAAGGUUCC-AAAAAUUACGAAUCGUCCGAUUCUUUUAC----CACACACACAC-ACGACUUGUGCGUGUGUG----UGUGCGGG- .............-.........(((((....))))).....(----(.(((((((((-(((.(....)))))))))----)))).)).- ( -30.10, z-score = -3.76, R) >droSim1.chr2R 9505199 82 - 19596830 UUUGAAAGGUUCCAAAAAAUUACGAAUCGUCCGAUUCUUUAAC--CACACACACACAC-ACGACUUGUACGUGUGUG----UGUGCGGG- ((((........)))).......(((((....))))).....(--(.(.(((((((((-(((.......))))))))----)))).)))- ( -27.60, z-score = -3.18, R) >droSec1.super_1 8258092 84 - 14215200 UUCGCAAGGUUCCAAAAAAUUACGAAUCGUCCGAUUCUUUAACCACACACACACACAC-ACGACUUGUACGUGUGUG----UGUGCGGG- .((....))..............(((((....))))).....((.(((((((((((((-(.....)))..)))))))----)))).)).- ( -28.70, z-score = -3.44, R) >droYak2.chr2R 10673119 83 - 21139217 UUUGCAAGGUUCCAAAAACUUAUGAAUCGUUCGAUUCUUUAAU--CACACACACACUCGACAACUUGUGUGUGUAUG----UGUGCGGG- .((((((((((.....)))))..(((((....)))))......--((((.((((((.(((....))).)))))).))----))))))).- ( -22.50, z-score = -1.97, R) >droEre2.scaffold_4845 7482137 78 + 22589142 UGUGCAAGGUUCCAAAAACUUACGAAUCGUUCGAUUCUUUAAU--CACACACACA-----CGACUCGUGUGUGUAUG----UGUGCAGG- ((..(((((((.....)))))..(((((....)))))......--((.(((((((-----((...))))))))).))----))..))..- ( -23.40, z-score = -2.11, R) >droAna3.scaffold_13266 8986350 87 - 19884421 -UUGGGAG--UCUAAUAAACAGUGGAAUGGGGGGUUUUUUGGUCUUCCAUAAGCCCAUACCUGUGUGUAUGUGUUUGUUCUUGUGUGGGG -.((((((--.((((.((((.............)))).)))).))))))....(((((((..(..((........))..)..))))))). ( -21.42, z-score = -0.23, R) >consensus UUUGCAAGGUUCCAAAAAAUUACGAAUCGUCCGAUUCUUUAAC__CACACACACACAC_ACGACUUGUACGUGUGUG____UGUGCGGG_ .(((((.................(((((....)))))............(((((((..............)))))))......))))).. ( -9.97 = -9.89 + -0.08)
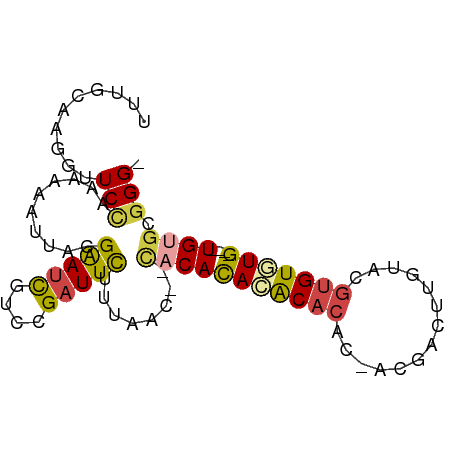
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:23:30 2011