| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,704,334 – 10,704,425 |
| Length | 91 |
| Max. P | 0.905800 |

| Location | 10,704,334 – 10,704,425 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 78.52 |
| Shannon entropy | 0.42460 |
| G+C content | 0.43327 |
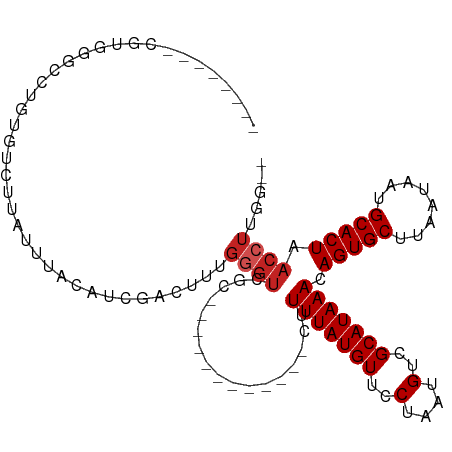
| Mean single sequence MFE | -23.75 |
| Consensus MFE | -12.19 |
| Energy contribution | -12.65 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
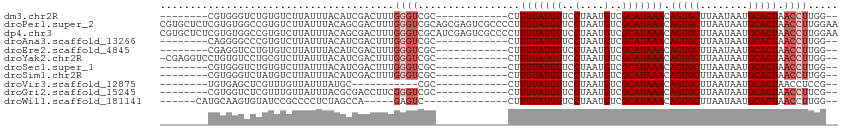
>dm3.chr2R 10704334 91 + 21146708 --------CGUGGGUCUGUGUCUUAUUUACAUCGACUUUGGGUCGC------------CUUUUAUGUUCCUAAUGUCGCAUAAACAGUGCUUAAUAAUGCACUAACCUUGG-- --------...((((..((((.(((((......(((.(((((..((------------.......)).))))).)))((((.....))))..))))).))))..))))...-- ( -22.20, z-score = -2.08, R) >droPer1.super_2 5119882 113 + 9036312 CGUGCUCUCGUGUGGCCGUGUCUUAUUUACAGCGACUUUGGGUCGCAGCGAGUCGCCCCUUUUAUGUUCCUAAUGUCGCAUAAACAGUGCUUAAUAAUGCACUAACCUUGGAA .(((..(((((((((((..(((...........)))....))))))).)))).))).((.(((((((..(....)..))))))).(((((........)))))......)).. ( -31.90, z-score = -1.36, R) >dp4.chr3 4927160 113 + 19779522 CGUGCUCUCGUGUGGCCGUGUCUUAUUUACAGCGACUUUGGGUCGCAUCGAGUCGCCCCUUUUAUGUUCCUAAUGUCGCAUAAACAGUGCUUAAUAAUGCACUAACCUUGGAA .(((..(((((((((((..(((...........)))....))))))).)))).))).((.(((((((..(....)..))))))).(((((........)))))......)).. ( -31.90, z-score = -1.91, R) >droAna3.scaffold_13266 8955710 91 + 19884421 --------CAGGGGCCCGUGUCUUAUUUACAUCGACUUUGGGUCGC------------CUUUUAUGUUCCUAAUGUCGCAUAAACAGUGCUUAAUAAUGCACUAACCUUGG-- --------.(((((((((.(((...........)))..)))))).)------------))(((((((..(....)..))))))).(((((........)))))........-- ( -25.40, z-score = -2.39, R) >droEre2.scaffold_4845 7449255 91 - 22589142 --------CGAGGUCCUGUGUCUUAUUUACAUCGACUUUGGGUCGC------------CUUUUAUGUUCCUAAUGUCGCAUAAACAGUGCUUAAUAAUGCACUAACCUUGG-- --------((((((...((((.(((((......(((.(((((..((------------.......)).))))).)))((((.....))))..))))).))))..)))))).-- ( -24.80, z-score = -3.24, R) >droYak2.chr2R 10640784 98 + 21139217 -CGAGGUCCUGUGUCCUGCGUCUUAUUUACAUCGACUUUGGGUCGC------------CUUUUAUGUUCCUAAUGUCGCAUAAACAGUGCUUAAUAAUGCACUAACCUUGG-- -((((((...(((.(((..(((...........)))...))).)))------------..(((((((..(....)..))))))).(((((........))))).)))))).-- ( -24.90, z-score = -2.35, R) >droSec1.super_1 8227149 91 + 14215200 --------CGUGGGUCUGUGUCUUAUUUACAUCGACUUUGGGUCGC------------CUUUUAUGUUCCUAAUGUCGCAUAAACAGUGCUUAAUAAUGCACUAACCUUGG-- --------...((((..((((.(((((......(((.(((((..((------------.......)).))))).)))((((.....))))..))))).))))..))))...-- ( -22.20, z-score = -2.08, R) >droSim1.chr2R 9473881 91 + 19596830 --------CGUGGGUCUAUGUCUUAUUUACAUCGACUUUGGGUCGC------------CUUUUAUGUUCCUAAUGUCGCAUAAACAGUGCUUAAUAAUGCACUAACCUUGG-- --------((.((((.(((((..((((.((((((((.....)))).------------.....))))....))))..))))).))(((((........)))))..)).)).-- ( -19.70, z-score = -1.44, R) >droVir3.scaffold_12875 7945669 80 - 20611582 --------UGUGAGCUCGUUUGUUAUUUAUGC-----------CGC------------CUUUUAUGUUCCUAAUGUCGCAUAAACAGUGCUUAAUAAUGCACUAACCUCCG-- --------.(.(((...........(((((((-----------.((------------...(((......))).)).))))))).(((((........)))))...)))).-- ( -14.30, z-score = -1.54, R) >droGri2.scaffold_15245 7693075 91 + 18325388 --------CGUGGUCUCGUUUGUUAUUUACGCGACCUUCGGGUCGC------------CUUUUAUGUUCCUAAUGUCGCAUAAACAGUGCUUAAUAAUGCACUAACCUUCG-- --------...(((................((((((....))))))------------..(((((((..(....)..))))))).(((((........))))).)))....-- ( -26.80, z-score = -4.08, R) >droWil1.scaffold_181141 942015 86 - 5303230 ------CAUGCAAGUGUAUCCGCCCCUCUAGCCA-----GAGUC--------------CUUUUAUGUUCCUAAUGUCGCAUAAACAGUGCUUAAUAAUGCACUAACCUUGG-- ------....((((.((........((((....)-----)))..--------------..(((((((..(....)..))))))).(((((........))))).)))))).-- ( -17.20, z-score = -1.63, R) >consensus ________CGUGGGCCUGUGUCUUAUUUACAUCGACUUUGGGUCGC____________CUUUUAUGUUCCUAAUGUCGCAUAAACAGUGCUUAAUAAUGCACUAACCUUGG__ .......................................((((.................(((((((..(....)..))))))).(((((........))))).))))..... (-12.19 = -12.65 + 0.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:23:25 2011