| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,702,700 – 10,702,791 |
| Length | 91 |
| Max. P | 0.520530 |

| Location | 10,702,700 – 10,702,791 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.08 |
| Shannon entropy | 0.46483 |
| G+C content | 0.40033 |
| Mean single sequence MFE | -18.78 |
| Consensus MFE | -11.42 |
| Energy contribution | -12.73 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.520530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
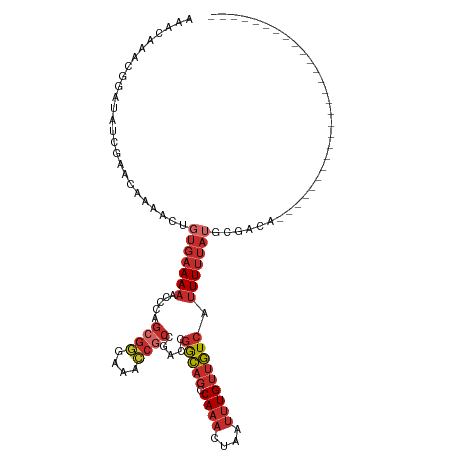
>dm3.chr2R 10702700 91 + 21146708 AAACAAACGGAUAUCGAACAAAACUGUGAAAAACCCAGCGGGAAACCGCCGACCGGCAGCCAAACUAAUUUGUUGUCAUUUUUAAGCGACA--------------------------- ........((...(((.(((....)))..........((((....)))))))))((((((.(((....)))))))))..............--------------------------- ( -18.60, z-score = -0.63, R) >droSim1.chr2R 9472304 91 + 19596830 AAACAAACGGAUAUCGAACAAAACUGUGAAAAACCCAGCGGGAAACCGCCGACCGGCAGCCAAACUAAUUUGUUGUCAUUUUUAUGCGACA--------------------------- .......((.((((((.(((....)))..........((((....)))))))..((((((.(((....))))))))).....))).))...--------------------------- ( -20.40, z-score = -1.17, R) >droSec1.super_1 8225595 91 + 14215200 AAACAAACGGAUAUCGAACAAAACUGUGAAAAACCCAGCGGGAAACCGCCGACCGGCAGCCAAACUAAUUUGUUGUCAUUUUUAUGCGACA--------------------------- .......((.((((((.(((....)))..........((((....)))))))..((((((.(((....))))))))).....))).))...--------------------------- ( -20.40, z-score = -1.17, R) >droYak2.chr2R 10639207 91 + 21139217 AAACAAACGGAUAUCGAACAAAACUGUGAAAAACCCAGCGUGAAACCGCCGACCGGCAGCCAAACUAAUUUGUUGUCAUUUUUAUGCGACA--------------------------- ........((.......(((....))).......)).((((((((..(((....))).....(((......))).....))))))))....--------------------------- ( -15.14, z-score = 0.23, R) >droEre2.scaffold_4845 7447653 91 - 22589142 AAACAAACGGAUAUCGAACAAAACUGUGAAAAACCCAGCGGGAAACCGCCGACCGGCAGCCAAACUAAUUUGUUGUCAUUUUUAUGCGACA--------------------------- .......((.((((((.(((....)))..........((((....)))))))..((((((.(((....))))))))).....))).))...--------------------------- ( -20.40, z-score = -1.17, R) >droAna3.scaffold_13266 8952034 90 + 19884421 AUACAAACGGAUAUCGAACAAAACUGUGAAAAACUCAGCGGGAAACCACCGAC-GGCAGUCAAACUAAUUUGUUGUCAUUUUUAUGCGACA--------------------------- .......((.(((..........(((.(.....).))).((....))...(((-(((((..........)))))))).....))).))...--------------------------- ( -17.00, z-score = -0.53, R) >droWil1.scaffold_181141 936845 78 - 5303230 -----AACGAA-----AACAAAACUGUGAAAAACUCAGCGGGAAACCGCG---UGGCAGUCAAACUAAUUUGUUGUCAUUUUUAUGCGACA--------------------------- -----......-----.........(((((((.....((((....)))).---((((((.((((....)))))))))))))))))......--------------------------- ( -19.80, z-score = -1.79, R) >droGri2.scaffold_15245 7691280 90 + 18325388 AAAAAUAUAUAUAUAUAACAGGACUGUGAAAAACACAGCGGGAAACCGCUCGC-GGCAGUCAAAUUAAUUUGUUGUCAUUUUUAUGCGACA--------------------------- .....................((((((......(.((((((....))))).).-))))))).........(((((.(((....))))))))--------------------------- ( -22.50, z-score = -2.80, R) >triCas2.ChLG4 8744239 114 - 13894384 AAAAACUCAAAUAUUGCGCAAAAUUUCUUAAAAAGAGACAAUAAAUCAGAGAACUAU-GUCAAA---AAAUGGCAUUAAUUUUACAUGUUCUGAGGUUUUAAACACGUUUUUAAACCA ...........(((((.......(((((.....))))))))))..((((((....((-((((..---...))))))............))))))(((((.(((....)))..))))). ( -14.80, z-score = -0.22, R) >consensus AAACAAACGGAUAUCGAACAAAACUGUGAAAAACCCAGCGGGAAACCGCCGACCGGCAGCCAAACUAAUUUGUUGUCAUUUUUAUGCGACA___________________________ .........................(((((((.....((((....)))).....(((((.((((....))))))))).)))))))................................. (-11.42 = -12.73 + 1.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:23:25 2011