
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,686,828 – 10,686,920 |
| Length | 92 |
| Max. P | 0.866060 |

| Location | 10,686,828 – 10,686,920 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 69.42 |
| Shannon entropy | 0.59476 |
| G+C content | 0.55296 |
| Mean single sequence MFE | -32.33 |
| Consensus MFE | -18.55 |
| Energy contribution | -18.17 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.77 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.866060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
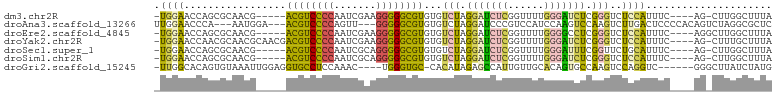
>dm3.chr2R 10686828 92 + 21146708 -UGGAACCAGCGCAACG-----ACGUCCCCAAUCGAAGGGGGCGUGUGUCUAGGAUCUCGGUUUUGGGAUCUCGGGUCUCCAUUUC----AG-CUUGGCUUUA -..(((((((.((.((.-----((((((((.......)))))))))).((..((((((((....))))))))..))..........----.)-))))).))). ( -31.60, z-score = -0.62, R) >droAna3.scaffold_13266 8935897 94 + 19884421 UUGGAACCCA---AAUGGA---ACGUCCCCAGUU---GGGGGCGUGUGUCUAGGAUCCCGUCCAUCCAAGUCCAAGUCUUGACUCCCCACAGUCUAGGCGCUC (((((.....---..((((---((((((((....---)))))))).......((((...)))).))))..)))))((((.((((......)))).)))).... ( -33.90, z-score = -2.08, R) >droEre2.scaffold_4845 7431355 93 - 22589142 -UGGAACCAGCGCAACG-----ACGUCCCCAAUCGAAGGGGGCGUGUGUCUAGGAUCUCGGUUUUGGGGCCUCGGGUCUCCAUUUC----AGGCUUGGCUUUA -.(((.(((((((....-----((((((((.......))))))))))).)).)).)))......(((((((...(((((.......----))))).))))))) ( -30.60, z-score = 0.49, R) >droYak2.chr2R 10622957 97 + 21139217 -UGGAACCAACGCAACGCAACGACGUCCCCAAUCGAAGGGGGCGUGUGUCUAGGAUCUCGGUUUUGGGAUCUCGGGUCUCCAUUUC----AG-CUUUGCUUUA -..........((((.((.((.((((((((.......)))))))))).((..((((((((....))))))))..))..........----.)-).)))).... ( -32.90, z-score = -1.19, R) >droSec1.super_1 8204908 92 + 14215200 -UGGAACCAGCGCAACG-----ACGUCCCCAAUCGCAGGGGGCGUGUGUCUAGGAUCUCGGUUUUGGGAUUUCGGUUCUGCAUUUC----AG-CUUGGCUUUA -.((((((.((((....-----((((((((.......))))))))))))...((((((((....)))))))).))))))((.....----.)-)......... ( -30.30, z-score = -0.64, R) >droSim1.chr2R 9456271 92 + 19596830 -UGGAACCAGCGCAACG-----ACGUCCCCAAUCGCAGGGGGCGUGUGUCUAGGAUCUCGGUUUUGGGAUCUCGGGUCUCCAUUUC----AG-CUUGGCUUUA -..(((((((.((.((.-----((((((((.......)))))))))).((..((((((((....))))))))..))..........----.)-))))).))). ( -31.60, z-score = -0.60, R) >droGri2.scaffold_15245 7670594 91 + 18325388 -UUGGCACAGUGUAAAUUGGAGGUGCCUCCAAAC----UGGGUGC-CACAUAGAGCCAUUGUUGCACAGUGCCAAGUCCAGGUC------GGGCUUAUCUAUG -.((((((...((...((((((....))))))))----...))))-))(((((((.(((((.....))))).)((((((.....------)))))).)))))) ( -35.40, z-score = -2.31, R) >consensus _UGGAACCAGCGCAACG_____ACGUCCCCAAUCGAAGGGGGCGUGUGUCUAGGAUCUCGGUUUUGGGAUCUCGGGUCUCCAUUUC____AG_CUUGGCUUUA .((((.................((((((((.......))))))))...(((.(((((((......))))))).)))..))))..................... (-18.55 = -18.17 + -0.37)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:23:24 2011