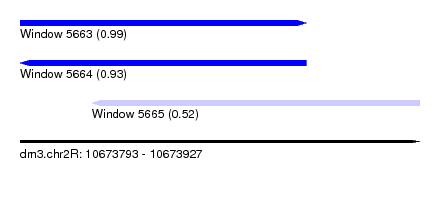
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,673,793 – 10,673,927 |
| Length | 134 |
| Max. P | 0.986228 |

| Location | 10,673,793 – 10,673,889 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 63.98 |
| Shannon entropy | 0.63812 |
| G+C content | 0.43788 |
| Mean single sequence MFE | -25.11 |
| Consensus MFE | -9.28 |
| Energy contribution | -9.41 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.986228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
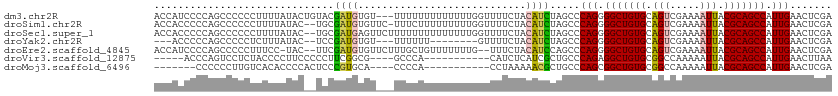
>dm3.chr2R 10673793 96 + 21146708 AUAUACGCACUCCAUAUAUCUAUAUC----------GAGUUCAAUGGCUGCGUAAUUUUC----GACUGCACAGCCCCUGGGCUAGAUGUAGAAAACCAAAAAAAAAAAA--- ..................((((((((----------.((((((..(((((.(((......----...))).)))))..)))))).)))))))).................--- ( -26.90, z-score = -4.22, R) >droSim1.chr2R 9442964 97 + 19596830 AUAUACGCACUC-AUAUAUCUAUAUC----------GAGUUCAAUGGCUGCGUAAUUUUC----GACUGCACAGCCCCUGGGCUAGAUGUAGAAAACCAAAAAAAAAGAAAG- ............-.....((((((((----------.((((((..(((((.(((......----...))).)))))..)))))).))))))))...................- ( -26.90, z-score = -3.93, R) >droSec1.super_1 8191910 98 + 14215200 AUAUACGCACUC-AUAUAUCUAUAUC----------GAGUUCAAUGGCUGCGUAAUUUUC----GACUGCACAGCCCCUGGGCUAGAUGUAGAAAACCAAAAAAAAAAAAAAG ............-.....((((((((----------.((((((..(((((.(((......----...))).)))))..)))))).)))))))).................... ( -26.90, z-score = -4.42, R) >droYak2.chr2R 10609682 87 + 21139217 GUAUACGCACUC-GUAUAUCUAUAUC----------GAGUUCAAUGGCUGCGUAAUUUUC----GACUGCACAGCCCCUGGGCUAGAUGUAGAAAACAAAAA----------- .((((((....)-)))))((((((((----------.((((((..(((((.(((......----...))).)))))..)))))).)))))))).........----------- ( -30.40, z-score = -4.66, R) >droEre2.scaffold_4845 7418390 106 - 22589142 GUAUACGCGCUC-AUAUAUCUAUAUCUCUACAUAUCGAGUUCAAUGGCUGCGUAAUUUUC----GACUGCACAGCCCCUGGGCUGGAUGUAGAAACAAAAAAACAGCAAAG-- ........(((.-((((....)))).((((((..((.((((((..(((((.(((......----...))).)))))..)))))).))))))))...........)))....-- ( -26.90, z-score = -2.16, R) >droVir3.scaffold_12875 7910557 75 - 20611582 ---------AUAUACGCACUCGGAUU----------AAGUUCAAUGGCUGCGUAAUUUUU----GGCCGCACAGCCUCUGGG-CAGCGAUGAGAUGUGG-------------- ---------.....((((((((....----------..(((((..(((((.((.......----....)).)))))..))))-).....)))).)))).-------------- ( -22.50, z-score = -0.81, R) >droMoj3.scaffold_6496 6884274 75 - 26866924 ---------ACGCACGCACUCGAAUC----------GAGUUCAAUGGCUGCGUAAUUUUU----GGCCGCACAGCCGCUGGG-CAGCGUUUUUAGGUGG-------------- ---------.......((((..((.(----------(.(((((.((((((.((.......----....)).)))))).))))-)..))..))..)))).-------------- ( -21.40, z-score = 0.57, R) >droGri2.scaffold_15245 7652142 88 + 18325388 ---------AUAAAUGCACACGCAUU----------GAGUUCAAUGGCUGCGUAAUUUUUUUUUGGUCGCACAGCCUCCGGGGCAGAAAUGAUAUUUAAGGGUAUGG------ ---------...(((((....)))))----------..((((...(((((.(..(((.......)))..).)))))....)))).......................------ ( -19.00, z-score = 0.36, R) >consensus _UAUACGCACUC_AUAUAUCUAUAUC__________GAGUUCAAUGGCUGCGUAAUUUUC____GACUGCACAGCCCCUGGGCUAGAUGUAGAAAACAAAAAAAA________ .......................................((((..(((((.((...............)).)))))..))))............................... ( -9.28 = -9.41 + 0.13)


| Location | 10,673,793 – 10,673,889 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 63.98 |
| Shannon entropy | 0.63812 |
| G+C content | 0.43788 |
| Mean single sequence MFE | -22.24 |
| Consensus MFE | -11.84 |
| Energy contribution | -11.73 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.930240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10673793 96 - 21146708 ---UUUUUUUUUUUUGGUUUUCUACAUCUAGCCCAGGGGCUGUGCAGUC----GAAAAUUACGCAGCCAUUGAACUC----------GAUAUAGAUAUAUGGAGUGCGUAUAU ---.............((((((.((...(((((....)))))....)).----))))))((((((.((((.....((----------......))...))))..))))))... ( -23.10, z-score = -1.13, R) >droSim1.chr2R 9442964 97 - 19596830 -CUUUCUUUUUUUUUGGUUUUCUACAUCUAGCCCAGGGGCUGUGCAGUC----GAAAAUUACGCAGCCAUUGAACUC----------GAUAUAGAUAUAU-GAGUGCGUAUAU -...................((((.(((.((..(((.(((((((.(((.----....))).))))))).)))..)).----------))).))))(((((-(....)))))). ( -22.50, z-score = -0.97, R) >droSec1.super_1 8191910 98 - 14215200 CUUUUUUUUUUUUUUGGUUUUCUACAUCUAGCCCAGGGGCUGUGCAGUC----GAAAAUUACGCAGCCAUUGAACUC----------GAUAUAGAUAUAU-GAGUGCGUAUAU ....................((((.(((.((..(((.(((((((.(((.----....))).))))))).)))..)).----------))).))))(((((-(....)))))). ( -22.50, z-score = -1.15, R) >droYak2.chr2R 10609682 87 - 21139217 -----------UUUUUGUUUUCUACAUCUAGCCCAGGGGCUGUGCAGUC----GAAAAUUACGCAGCCAUUGAACUC----------GAUAUAGAUAUAC-GAGUGCGUAUAC -----------.........((((.(((.((..(((.(((((((.(((.----....))).))))))).)))..)).----------))).))))(((((-(....)))))). ( -24.20, z-score = -2.19, R) >droEre2.scaffold_4845 7418390 106 + 22589142 --CUUUGCUGUUUUUUUGUUUCUACAUCCAGCCCAGGGGCUGUGCAGUC----GAAAAUUACGCAGCCAUUGAACUCGAUAUGUAGAGAUAUAGAUAUAU-GAGCGCGUAUAC --....(((((.(((.((((((((((((.((..(((.(((((((.(((.----....))).))))))).)))..)).))..)))))))))).)))...))-.)))........ ( -28.50, z-score = -1.51, R) >droVir3.scaffold_12875 7910557 75 + 20611582 --------------CCACAUCUCAUCGCUG-CCCAGAGGCUGUGCGGCC----AAAAAUUACGCAGCCAUUGAACUU----------AAUCCGAGUGCGUAUAU--------- --------------...........(((..-..(((.(((((((..(..----.....)..))))))).))).((((----------.....))))))).....--------- ( -16.80, z-score = -0.18, R) >droMoj3.scaffold_6496 6884274 75 + 26866924 --------------CCACCUAAAAACGCUG-CCCAGCGGCUGUGCGGCC----AAAAAUUACGCAGCCAUUGAACUC----------GAUUCGAGUGCGUGCGU--------- --------------..........((((((-(.(((.(((((((..(..----.....)..))))))).))).((((----------(...)))))))).))))--------- ( -23.90, z-score = -1.34, R) >droGri2.scaffold_15245 7652142 88 - 18325388 ------CCAUACCCUUAAAUAUCAUUUCUGCCCCGGAGGCUGUGCGACCAAAAAAAAAUUACGCAGCCAUUGAACUC----------AAUGCGUGUGCAUUUAU--------- ------.(((((..............((((...))))(((((((..(...........)..)))))))((((....)----------)))..))))).......--------- ( -16.40, z-score = -0.61, R) >consensus ________UUUUUUUGGUUUUCUACAUCUAGCCCAGGGGCUGUGCAGUC____GAAAAUUACGCAGCCAUUGAACUC__________GAUAUAGAUAUAU_GAGUGCGUAUA_ .................................(((.(((((((.................))))))).)))......................................... (-11.84 = -11.73 + -0.11)



| Location | 10,673,817 – 10,673,927 |
|---|---|
| Length | 110 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 69.34 |
| Shannon entropy | 0.57316 |
| G+C content | 0.50542 |
| Mean single sequence MFE | -20.71 |
| Consensus MFE | -12.73 |
| Energy contribution | -12.81 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -0.49 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.523730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10673817 110 - 21146708 ACCAUCCCCAGCCCCCCUUUUAUACUGUACGAUGUGU---UUUUUUUUUUUUUGGUUUUCUACAUCUAGCCCAGGGGCUGUGCAGUCGAAAAUUACGCAGCCAUUGAACUCGA ..........((.....((((..(((((((((((((.---....................)))))).((((....)))))))))))..))))....))............... ( -21.00, z-score = -0.21, R) >droSim1.chr2R 9442987 110 - 19596830 ACCACCCCCAGCCCCCCUUUUAUAC--UGCGAUGUGUUC-UUUCUUUUUUUUUGGUUUUCUACAUCUAGCCCAGGGGCUGUGCAGUCGAAAAUUACGCAGCCAUUGAACUCGA ..........((.....((((..((--(((((((((...-...((........)).....))))))(((((....))))).)))))..))))....))............... ( -20.70, z-score = 0.10, R) >droSec1.super_1 8191933 111 - 14215200 ACCACCCCCAGCCCCCCUUUUAUAC--UGCGAUGAGUUCUUUUUUUUUUUUUUGGUUUUCUACAUCUAGCCCAGGGGCUGUGCAGUCGAAAAUUACGCAGCCAUUGAACUCGA .........................--.....(((((((...........(((((....(((....))).)))))(((((((.(((.....))).)))))))...))))))). ( -22.30, z-score = -0.57, R) >droYak2.chr2R 10609705 97 - 21139217 ---ACCCCCAGCCCCCUCUUUAUAC--UCCGAUGUGU---UUUUUU--------GUUUUCUACAUCUAGCCCAGGGGCUGUGCAGUCGAAAAUUACGCAGCCAUUGAACUCGA ---......................--..(((((.((---(.....--------((((((.((...(((((....)))))....)).)))))).....))))))))....... ( -18.00, z-score = -0.12, R) >droEre2.scaffold_4845 7418423 108 + 22589142 ACCAUCCCCAGCCCCCUUUCC-UAC--UUCGAUGUGUUCUUUGCUGUUUUUUUG--UUUCUACAUCCAGCCCAGGGGCUGUGCAGUCGAAAAUUACGCAGCCAUUGAACUCGA .....................-...--.((((...((((..((((((.((((((--...((.((..(((((....))))))).)).))))))....)))).))..)))))))) ( -25.10, z-score = -1.68, R) >droVir3.scaffold_12875 7910572 93 + 20611582 -----ACCCAGUCCUCUACCCCUUCCCCCUUCGGCG----GCCCA-----------CAUCUCAUCGCUGCCCAGAGGCUGUGCGGCCAAAAAUUACGCAGCCAUUGAACUUAA -----...........................((((----((...-----------.........))))))(((.(((((((..(.......)..))))))).)))....... ( -21.80, z-score = -0.93, R) >droMoj3.scaffold_6496 6884289 91 + 26866924 -------CCCCCCUUGUCACACCCCACUCCCGUGCA----CCCCA-----------CCUAAAAACGCUGCCCAGCGGCUGUGCGGCCAAAAAUUACGCAGCCAUUGAACUCGA -------.......((.(((...........)))))----.....-----------.........(((....)))(((((((..(.......)..)))))))........... ( -16.10, z-score = -0.06, R) >consensus ACCACCCCCAGCCCCCUUUUUAUAC__UCCGAUGUGU___UUUCUUUUUUUUUGGUUUUCUACAUCUAGCCCAGGGGCUGUGCAGUCGAAAAUUACGCAGCCAUUGAACUCGA .......................................................................(((.(((((((.(((.....))).))))))).)))....... (-12.73 = -12.81 + 0.08)
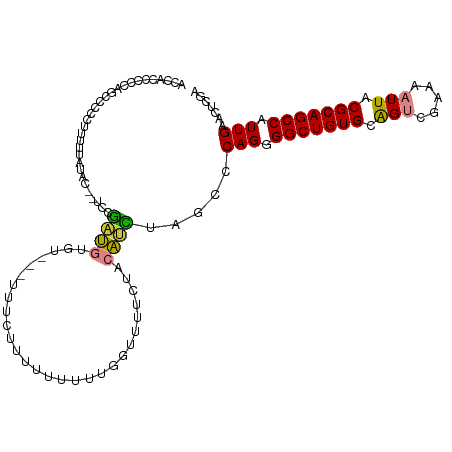
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:23:21 2011