| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,658,335 – 10,658,427 |
| Length | 92 |
| Max. P | 0.716633 |

| Location | 10,658,335 – 10,658,427 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 69.49 |
| Shannon entropy | 0.61175 |
| G+C content | 0.53843 |
| Mean single sequence MFE | -24.75 |
| Consensus MFE | -11.23 |
| Energy contribution | -11.69 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.516001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
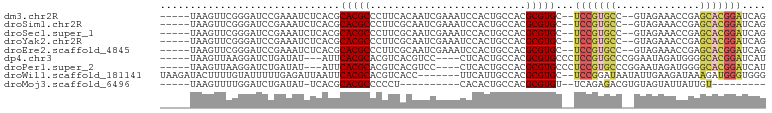
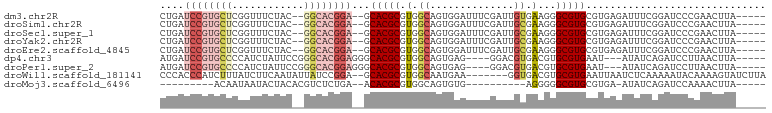
>dm3.chr2R 10658335 92 + 21146708 -----UAAGUUCGGGAUCCGAAAUCUCACGCACGCCCUUCACAAUCGAAAUCCACUGCCACGCGUGC--UCCGUGCC--GUAGAAACCGAGCACGGAUCAG -----....(((((...))))).......((((((..........................))))))--((((((((--(.......)).))))))).... ( -25.17, z-score = -1.26, R) >droSim1.chr2R 9427599 92 + 19596830 -----UAAGUUCGGGAUCCGAAAUCUCACGCACGCCCUUCGCAAUCGAAAUCCACUGCCACGCGUGC--UCCGUGCC--GUAGAAACCGAGCACGGAUCAG -----....(((((...))))).......((((((.....(((............)))...))))))--((((((((--(.......)).))))))).... ( -27.80, z-score = -1.59, R) >droSec1.super_1 8176513 92 + 14215200 -----UAAGUUCGGGAUCCGAAAUCUCACGCACGCCCUUCGCAAUCGAAAUCCACUGCCACGCGUGC--UCCGUGCC--GUAGAAACCGAGCACGGAUCAG -----....(((((...))))).......((((((.....(((............)))...))))))--((((((((--(.......)).))))))).... ( -27.80, z-score = -1.59, R) >droYak2.chr2R 10593848 92 + 21139217 -----UAAGUUCGGGAUCCGAAAUCUCACGCACGCCCUUCGCAAUCGAAAUCCACUGCCACGCGUGC--UCCGUGCC--GUAGAAACCGAGCACGGAUCAG -----....(((((...))))).......((((((.....(((............)))...))))))--((((((((--(.......)).))))))).... ( -27.80, z-score = -1.59, R) >droEre2.scaffold_4845 7402910 92 - 22589142 -----UAAGUUCGGGAUCCGAAAUCUCACGCACGCCCUUCGCAAUCGAAAUCCACUGCCACGCGUGC--UCCGUGCC--GUAGAAACCGAGCACGGAUCAG -----....(((((...))))).......((((((.....(((............)))...))))))--((((((((--(.......)).))))))).... ( -27.80, z-score = -1.59, R) >dp4.chr3 4877003 89 + 19779522 -----UAAGUUAAGGAUCUGAUAU---AUUCACGCACGUCACGUCC----CUCACUGCCACGCGUGCCCUCCGUGCCCGGAAUAGAUGGGGCACGGAUCAU -----........((...(((...---..))).((((((...((..----......))...))))))))(((((((((.(......).))))))))).... ( -27.50, z-score = -0.85, R) >droPer1.super_2 5069485 89 + 9036312 -----UAAGUUAAGGAUCUGAUAU---AUUCACGCACGUCACGUCC----CUCACUGCCACGCGUGCCCUCCGUGCCCGGAAUAGAUGGGGCACGGAUCAU -----........((...(((...---..))).((((((...((..----......))...))))))))(((((((((.(......).))))))))).... ( -27.50, z-score = -0.85, R) >droWil1.scaffold_181141 865549 92 - 5303230 UAAGAUACUUUUGUAUUUUUGAGAUUAAUUCACGCACGUCACC-------UUCAUUGCCACGCGUGC--UCCGGAUAAUAUUGAAGAUAAAGAUGGGUGGG .....(((((...((((((..(.((((.(((..((((((...(-------......)....))))))--...))))))).)..)))))).....))))).. ( -16.80, z-score = 0.48, R) >droMoj3.scaffold_6496 6862860 74 - 26866924 -----UAAGUUUUGGAUCUGAUAU-UCACGCACGCCCCCU----------CACACUGCCACGCGUGU--UCAGAGACGUGUAGUAUUAUUGU--------- -----........(..(((((...-.(((((..((.....----------......))...))))).--)))))..)...............--------- ( -14.60, z-score = 0.06, R) >consensus _____UAAGUUCGGGAUCCGAAAUCUCACGCACGCCCUUCACAAUCGAAAUCCACUGCCACGCGUGC__UCCGUGCC__GUAGAAACCGAGCACGGAUCAG ..............................(((((..........................)))))...(((((((..............))))))).... (-11.23 = -11.69 + 0.46)
| Location | 10,658,335 – 10,658,427 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 69.49 |
| Shannon entropy | 0.61175 |
| G+C content | 0.53843 |
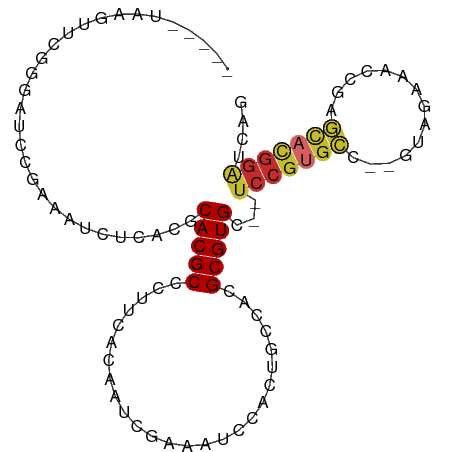
| Mean single sequence MFE | -31.42 |
| Consensus MFE | -13.20 |
| Energy contribution | -14.27 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.716633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
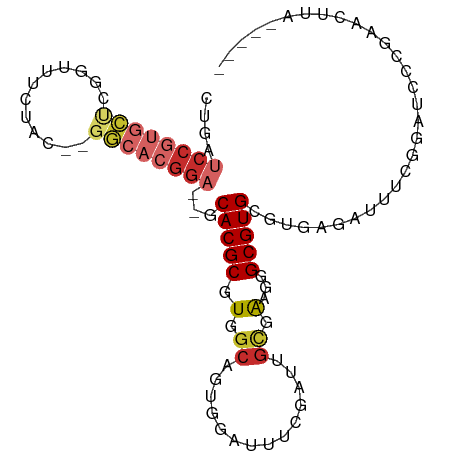
>dm3.chr2R 10658335 92 - 21146708 CUGAUCCGUGCUCGGUUUCUAC--GGCACGGA--GCACGCGUGGCAGUGGAUUUCGAUUGUGAAGGGCGUGCGUGAGAUUUCGGAUCCCGAACUUA----- ....(((((((.((.......)--))))))))--((((((.(.(((((.(....).))))).)...)))))).((((..(((((...)))))))))----- ( -36.50, z-score = -1.90, R) >droSim1.chr2R 9427599 92 - 19596830 CUGAUCCGUGCUCGGUUUCUAC--GGCACGGA--GCACGCGUGGCAGUGGAUUUCGAUUGCGAAGGGCGUGCGUGAGAUUUCGGAUCCCGAACUUA----- ....(((((((.((.......)--))))))))--((((((.(.(((((.(....).)))))...).)))))).((((..(((((...)))))))))----- ( -38.00, z-score = -2.08, R) >droSec1.super_1 8176513 92 - 14215200 CUGAUCCGUGCUCGGUUUCUAC--GGCACGGA--GCACGCGUGGCAGUGGAUUUCGAUUGCGAAGGGCGUGCGUGAGAUUUCGGAUCCCGAACUUA----- ....(((((((.((.......)--))))))))--((((((.(.(((((.(....).)))))...).)))))).((((..(((((...)))))))))----- ( -38.00, z-score = -2.08, R) >droYak2.chr2R 10593848 92 - 21139217 CUGAUCCGUGCUCGGUUUCUAC--GGCACGGA--GCACGCGUGGCAGUGGAUUUCGAUUGCGAAGGGCGUGCGUGAGAUUUCGGAUCCCGAACUUA----- ....(((((((.((.......)--))))))))--((((((.(.(((((.(....).)))))...).)))))).((((..(((((...)))))))))----- ( -38.00, z-score = -2.08, R) >droEre2.scaffold_4845 7402910 92 + 22589142 CUGAUCCGUGCUCGGUUUCUAC--GGCACGGA--GCACGCGUGGCAGUGGAUUUCGAUUGCGAAGGGCGUGCGUGAGAUUUCGGAUCCCGAACUUA----- ....(((((((.((.......)--))))))))--((((((.(.(((((.(....).)))))...).)))))).((((..(((((...)))))))))----- ( -38.00, z-score = -2.08, R) >dp4.chr3 4877003 89 - 19779522 AUGAUCCGUGCCCCAUCUAUUCCGGGCACGGAGGGCACGCGUGGCAGUGAG----GGACGUGACGUGCGUGAAU---AUAUCAGAUCCUUAACUUA----- .((((((((((((..........)))))))))..((.(((((.((.((...----..)))).))))).))....---...))).............----- ( -29.10, z-score = -0.30, R) >droPer1.super_2 5069485 89 - 9036312 AUGAUCCGUGCCCCAUCUAUUCCGGGCACGGAGGGCACGCGUGGCAGUGAG----GGACGUGACGUGCGUGAAU---AUAUCAGAUCCUUAACUUA----- .((((((((((((..........)))))))))..((.(((((.((.((...----..)))).))))).))....---...))).............----- ( -29.10, z-score = -0.30, R) >droWil1.scaffold_181141 865549 92 + 5303230 CCCACCCAUCUUUAUCUUCAAUAUUAUCCGGA--GCACGCGUGGCAAUGAA-------GGUGACGUGCGUGAAUUAAUCUCAAAAAUACAAAAGUAUCUUA ..(((.(((..(((((((((.(.((((.((..--...)).)))).).))))-------))))).))).)))..............((((....)))).... ( -15.70, z-score = -0.14, R) >droMoj3.scaffold_6496 6862860 74 + 26866924 ---------ACAAUAAUACUACACGUCUCUGA--ACACGCGUGGCAGUGUG----------AGGGGGCGUGCGUGA-AUAUCAGAUCCAAAACUUA----- ---------.......(((..(((((((((..--.(((((......)))))----------.))))))))).))).-...................----- ( -20.40, z-score = -1.19, R) >consensus CUGAUCCGUGCUCGGUUUCUAC__GGCACGGA__GCACGCGUGGCAGUGGAUUUCGAUUGCGAAGGGCGUGCGUGAGAUUUCGGAUCCCGAACUUA_____ ....((((((((............))))))))...(((((.(.((..............)).)...))))).............................. (-13.20 = -14.27 + 1.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:23:18 2011