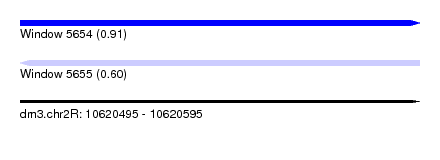
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,620,495 – 10,620,595 |
| Length | 100 |
| Max. P | 0.912619 |

| Location | 10,620,495 – 10,620,595 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.56 |
| Shannon entropy | 0.40564 |
| G+C content | 0.42415 |
| Mean single sequence MFE | -26.82 |
| Consensus MFE | -15.74 |
| Energy contribution | -16.38 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.912619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10620495 100 + 21146708 -AGUCCGAGACAUCGUCGAAACUUUUGCCAUAGCUC--GAACAUAAUUAUAAAAGUUUCGACAAGUAUUUGCCCAGGUGGGCCA------------GUGUUCGUGCAAAAAAUAA--- -.(..((((.(((.(((((((((((((.........--...........)))))))))))))........((((....))))..------------)))))))..).........--- ( -27.75, z-score = -3.01, R) >droYak2.chr2R 10554366 112 + 21139217 -AGUCCGAGACAUCGUCGAAACUUUUGCCAUAGCUC--GAACAUAAUUAUAAAAGUUUCGACAAGUAUUUGCCCAGGUGGGCCAGCAACGCAGCCAGUGUUCGUGCAAAAAAUAA--- -.(..((((.(((.(((((((((((((.........--...........)))))))))))))........((((....))))..((......))..)))))))..).........--- ( -27.85, z-score = -1.63, R) >droEre2.scaffold_4845 7361930 109 - 22589142 -AGUCCGAGACAUCGUCGAAACUUUUGCCAUAGCUC--GAACAUAAUUAUAAAAGUUUCGACAAGUAUUUGCCCAGGUGGGCCAACA---CAGCCAGUGUUCGGGCAAAAAACAG--- -.(((((((.(((.(((((((((((((.........--...........)))))))))))))..((.((.((((....)))).)).)---).....)))))))))).........--- ( -34.55, z-score = -4.15, R) >droAna3.scaffold_13266 5100056 102 - 19884421 GAGUCCGAGACAUCGUCGGAACUUUUGCCAUAGCUC--GAACAUAAUUAUAAAAGUUUCGACAAGUAUUUGCCCAGGUGGGCUA--------------GCUCUUGCAAAAAACAAAAA ..((..(((.(...(((((((((((((.........--...........)))))))))))))........((((....))))..--------------))))..))............ ( -24.95, z-score = -1.65, R) >dp4.chr3 5280668 104 + 19779522 -AGUCCGAGACGUGGCUAAAACUUUUGCCAUUGCUC--GAACAUAAUUAUAAAAGUUUCGACAAGUAUUUGGCCAGGUGGGCAAGGA--------GCAGUUCCACACAGAAACAA--- -.((.((((..(((((..........)))))..)))--).))............(((((.........(((.((....)).)))(((--------(...)))).....)))))..--- ( -26.60, z-score = -1.35, R) >droPer1.super_2 5485945 104 + 9036312 -AGUCCGAGACGUGGCUAAAACUUUUGCCAUUGCUC--GAACAUAAUUAUAAAAGUUUCGACAAGUAUUUGGCCAGGUGGGCAAGGA--------GCAGUUCCAUACAGAAACAA--- -.((.((((..(((((..........)))))..)))--).))............(((((.........(((.((....)).)))(((--------(...)))).....)))))..--- ( -26.60, z-score = -1.43, R) >droWil1.scaffold_180699 1345704 89 + 2593675 -GGUCCGAGACAUCGUUGAAACUUUUGCCAUAGCUCCUGAACAUAAUUAUAAAAGUUUCGACAAGUAUUUGCC-AGGUGGGCAAAAAGCAA--------------------------- -.(((...)))...(((((((((((((......................))))))))))))).....((((((-.....))))))......--------------------------- ( -19.45, z-score = -0.79, R) >consensus _AGUCCGAGACAUCGUCGAAACUUUUGCCAUAGCUC__GAACAUAAUUAUAAAAGUUUCGACAAGUAUUUGCCCAGGUGGGCAAG_A_________GUGUUCGUGCAAAAAACAA___ ..(((...)))...(((((((((((((......................)))))))))))))........((((....)))).................................... (-15.74 = -16.38 + 0.63)

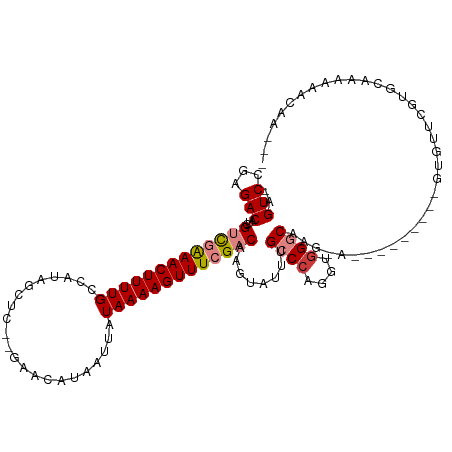
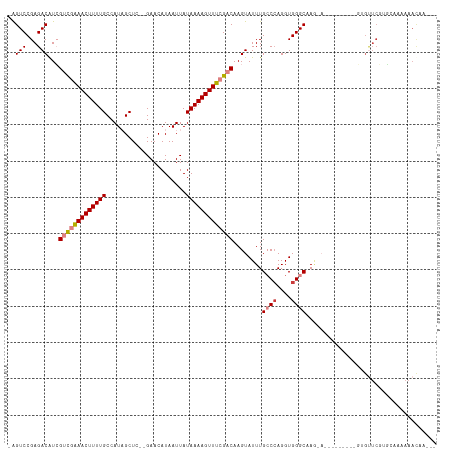
| Location | 10,620,495 – 10,620,595 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.56 |
| Shannon entropy | 0.40564 |
| G+C content | 0.42415 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -14.37 |
| Energy contribution | -15.03 |
| Covariance contribution | 0.65 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.597308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10620495 100 - 21146708 ---UUAUUUUUUGCACGAACAC------------UGGCCCACCUGGGCAAAUACUUGUCGAAACUUUUAUAAUUAUGUUC--GAGCUAUGGCAAAAGUUUCGACGAUGUCUCGGACU- ---.........(..((((((.------------..((((....))))......((((((((((((((...((...((..--..)).))...))))))))))))))))).)))..).- ( -27.20, z-score = -2.79, R) >droYak2.chr2R 10554366 112 - 21139217 ---UUAUUUUUUGCACGAACACUGGCUGCGUUGCUGGCCCACCUGGGCAAAUACUUGUCGAAACUUUUAUAAUUAUGUUC--GAGCUAUGGCAAAAGUUUCGACGAUGUCUCGGACU- ---.........(..((((((..(((......))).((((....))))......((((((((((((((...((...((..--..)).))...))))))))))))))))).)))..).- ( -29.30, z-score = -1.42, R) >droEre2.scaffold_4845 7361930 109 + 22589142 ---CUGUUUUUUGCCCGAACACUGGCUG---UGUUGGCCCACCUGGGCAAAUACUUGUCGAAACUUUUAUAAUUAUGUUC--GAGCUAUGGCAAAAGUUUCGACGAUGUCUCGGACU- ---...........(((((((......(---((((.((((....)))).)))))((((((((((((((...((...((..--..)).))...))))))))))))))))).))))...- ( -33.80, z-score = -2.89, R) >droAna3.scaffold_13266 5100056 102 + 19884421 UUUUUGUUUUUUGCAAGAGC--------------UAGCCCACCUGGGCAAAUACUUGUCGAAACUUUUAUAAUUAUGUUC--GAGCUAUGGCAAAAGUUCCGACGAUGUCUCGGACUC .......(((((((...(((--------------..((((....)))).........(((((.(............))))--)))))...))))))).(((((.(....))))))... ( -26.80, z-score = -1.88, R) >dp4.chr3 5280668 104 - 19779522 ---UUGUUUCUGUGUGGAACUGC--------UCCUUGCCCACCUGGCCAAAUACUUGUCGAAACUUUUAUAAUUAUGUUC--GAGCAAUGGCAAAAGUUUUAGCCACGUCUCGGACU- ---..(((((.(((((((.....--------)))..(((.....)))...)))).....)))))............((((--(((...((((..........))))...))))))).- ( -23.50, z-score = -0.74, R) >droPer1.super_2 5485945 104 - 9036312 ---UUGUUUCUGUAUGGAACUGC--------UCCUUGCCCACCUGGCCAAAUACUUGUCGAAACUUUUAUAAUUAUGUUC--GAGCAAUGGCAAAAGUUUUAGCCACGUCUCGGACU- ---..(((((.(((((((.....--------)))..(((.....)))...)))).....)))))............((((--(((...((((..........))))...))))))).- ( -23.50, z-score = -1.04, R) >droWil1.scaffold_180699 1345704 89 - 2593675 ---------------------------UUGCUUUUUGCCCACCU-GGCAAAUACUUGUCGAAACUUUUAUAAUUAUGUUCAGGAGCUAUGGCAAAAGUUUCAACGAUGUCUCGGACC- ---------------------------......((((((.....-)))))).....((((((((((((...((...(((....))).))...)))))))))..(((....)))))).- ( -18.60, z-score = -0.73, R) >consensus ___UUGUUUUUUGCACGAACAC_________U_CUGGCCCACCUGGGCAAAUACUUGUCGAAACUUUUAUAAUUAUGUUC__GAGCUAUGGCAAAAGUUUCGACGAUGUCUCGGACU_ ....................................((((....))))........((((((((((((...((...(((....))).))...)))))))))..(((....)))))).. (-14.37 = -15.03 + 0.65)
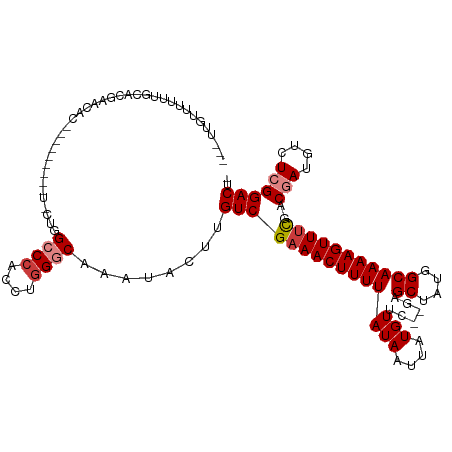
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:23:13 2011