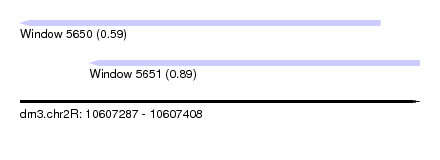
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,607,287 – 10,607,408 |
| Length | 121 |
| Max. P | 0.888777 |

| Location | 10,607,287 – 10,607,396 |
|---|---|
| Length | 109 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.21 |
| Shannon entropy | 0.57260 |
| G+C content | 0.41920 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -15.32 |
| Energy contribution | -16.27 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.45 |
| Mean z-score | -0.63 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.587378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
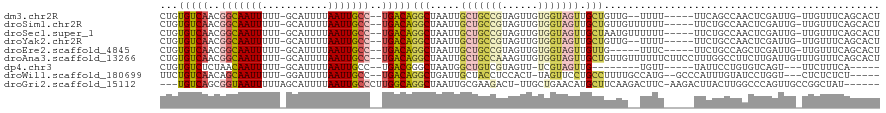
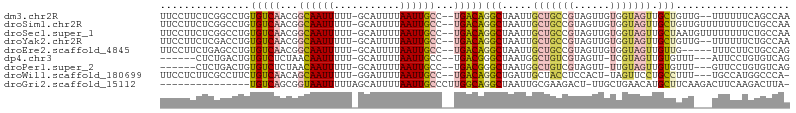
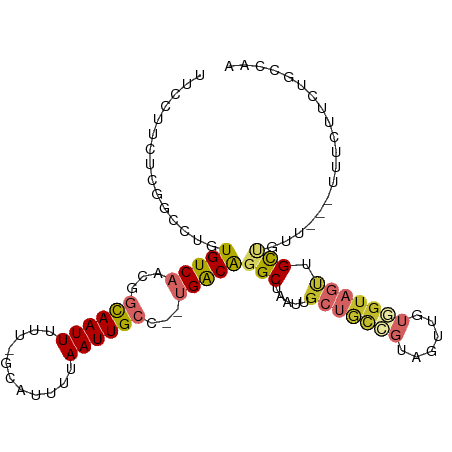
>dm3.chr2R 10607287 109 - 21146708 CUGUGUCAACGGCAAUUUUU-GCAUUUUAAUUGCC--UGACAGGCUAAUUGCUGCCGUAGUUGUGGUAGUUGCUGUUG--UUUU-----UUCAGCCAACUCGAUUG-UUGUUUCAGCACU ..(((((((((((((((.(.-.(.........(((--.....))).((((((....)))))))..).)))))))))))--....-----......((((......)-))).....)))). ( -29.70, z-score = -1.05, R) >droSim1.chr2R 9371267 111 - 19596830 CUGUGUCAACGGCAAUUUUU-GCAUUUUAAUUGCC--UGACAGGCUAAUUGCUGCCGUAGUUGUGGUAGUUGCUGUUGUUUUUU-----UUCUGCCAACUCGAUUG-UUGUUUCAGCACU ..(((((((((((((((.(.-.(.........(((--.....))).((((((....)))))))..).)))))))))))......-----......((((......)-))).....)))). ( -29.70, z-score = -1.32, R) >droSec1.super_1 8127650 111 - 14215200 CUGUGUCAACGGCAAUUUUU-GCAUUUUAAUUGCC--UGACAGGCUAAUUGCUGCCGUAGUUGUGGUAGUUGCUAAUGUUUUUU-----UUCUGCCAACUCGAUUG-UUGUUUCAGCACU ..((((.(((((((......-(((((.((((((((--((((.(((........)))...)))).))))))))..))))).....-----...))))(((......)-)))))...)))). ( -28.04, z-score = -0.97, R) >droYak2.chr2R 10540058 109 - 21139217 CUGUGUCAACGGCAAUUUUU-GCAUUUUAAUUGCC--UGACAGGCUAAUUGCUGCCGUAGUUGUGGUAGUUGCUGUUG--UUUU-----UUCUGCCAACUCGAUUG-UUGUUUCAGCACU ..(((((((((((((((.(.-.(.........(((--.....))).((((((....)))))))..).)))))))))))--....-----......((((......)-))).....)))). ( -29.70, z-score = -1.26, R) >droEre2.scaffold_4845 7349185 106 + 22589142 CUGUGUCAACGGCAAUUUUU-GCAUUUUAAUUGCC--UGACAGGCUAAUUGCUGCCGUAGUUGUGGUAGUUGUUG-----UUUC-----UUCUGCCAGCUCGAUUG-UUGUUUCAGCACU ..((((.((((((((((...-((.........(((--.....)))(((((((....)))))))((((((..(...-----...)-----..))))))))..)))))-)))))...)))). ( -29.20, z-score = -1.20, R) >droAna3.scaffold_13266 5088879 117 + 19884421 CUGUGUCAACGGCAAUUUUU-GCAUUUUAAUUGCC--UGACAGGCUAAUUGCUGCCAAAGUUGUGGUAGUUGCUGUUGUUUUUUCUUCCUUUGGCCUUCUUGAUUGUUUGUUUCAGCACU ..((((.((((((((((...-.......)))))))--.((.(((((((..(((((((......))))))).((....))...........)))))))))..........)))...)))). ( -27.70, z-score = -0.67, R) >dp4.chr3 5260207 95 - 19779522 CUGUGUCUCUAACAAUUUUU-GCAUUUUAAUUGCC--UGACGGGCUAAUGGCUGUCGUAGUU-UCGUAGUUG--------UGUU-----UAUUCCUGUGUCAGU---UUCUUUCA----- .(((.......)))......-(((.......)))(--(((((((...(..((((.((.....-.))))))..--------)...-----....)))..))))).---........----- ( -14.70, z-score = 0.16, R) >droWil1.scaffold_180699 1328327 106 - 2593675 UUCUGUCAACAGCAAUUUUU-GGAUUUUAAUUGCC--UGACAGGCUGAUUGCUACCUCCACU-UAGUUCCUGCCUUUUGCCAUG--GCCCAUUUGUAUCCUGGU---CUCUCUCU----- ...(((((...((((((...-.......)))))).--)))))(((.....((((........-))))....)))....((((.(--(..(....)..)).))))---........----- ( -18.60, z-score = 0.45, R) >droGri2.scaffold_15112 4638755 109 + 5172618 ---UGUCAGCGGUAAUUUUUAGCAUUUUAAUUGCCCUUGGCAGGCUAAUUGCGAAGACU-UUGCUGAACAUGCUUCAAGACUUC-AAGACUUACUUGGCCCAGUUGCCGGCUAU------ ---((((((((((((((...........)))))))(((.((((.....)))).)))...-..)))).))).(((....((((((-(((.....)))))...))))...)))...------ ( -26.70, z-score = 0.21, R) >consensus CUGUGUCAACGGCAAUUUUU_GCAUUUUAAUUGCC__UGACAGGCUAAUUGCUGCCGUAGUUGUGGUAGUUGCUGUUG__UUUU_____UUCUGCCAACUCGAUUG_UUGUUUCAGCACU ...(((((..(((((((...........)))))))..)))))(((.....(((((((......))))))).))).............................................. (-15.32 = -16.27 + 0.95)
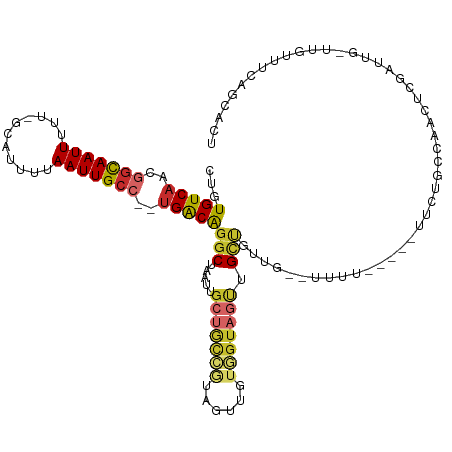
| Location | 10,607,308 – 10,607,408 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 72.81 |
| Shannon entropy | 0.56088 |
| G+C content | 0.42822 |
| Mean single sequence MFE | -24.43 |
| Consensus MFE | -11.56 |
| Energy contribution | -12.46 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.888777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10607308 100 - 21146708 UUCCUUCUCGGCCUGUGUCAACGGCAAUUUUU-GCAUUUUAAUUGCC--UGACAGGCUAAUUGCUGCCGUAGUUGUGGUAGUUGCUGUUG--UUUUUUCAGCCAA .........(((.((...(((((((((((.(.-.(.........(((--.....))).((((((....)))))))..).)))))))))))--......))))).. ( -29.60, z-score = -2.15, R) >droSim1.chr2R 9371288 102 - 19596830 UUCCUUCUCGGCCUGUGUCAACGGCAAUUUUU-GCAUUUUAAUUGCC--UGACAGGCUAAUUGCUGCCGUAGUUGUGGUAGUUGCUGUUGUUUUUUUUCUGCCAA .........(((...(((((..(((((((...-.......)))))))--)))))(((.....(((((((......))))))).)))..............))).. ( -27.40, z-score = -1.70, R) >droSec1.super_1 8127671 102 - 14215200 UUCCUUCUCGGCCUGUGUCAACGGCAAUUUUU-GCAUUUUAAUUGCC--UGACAGGCUAAUUGCUGCCGUAGUUGUGGUAGUUGCUAAUGUUUUUUUUCUGCCAA .........(((...(((((..(((((((...-.......)))))))--)))))(((.....(((((((......))))))).)))..............))).. ( -27.40, z-score = -1.83, R) >droYak2.chr2R 10540079 100 - 21139217 UUCCUUCUCGACCUGUGUCAACGGCAAUUUUU-GCAUUUUAAUUGCC--UGACAGGCUAAUUGCUGCCGUAGUUGUGGUAGUUGCUGUUG--UUUUUUCUGCCAA ........((((...(((((..(((((((...-.......)))))))--)))))(((........)))...))))((((((..((....)--).....)))))). ( -26.40, z-score = -1.87, R) >droEre2.scaffold_4845 7349206 97 + 22589142 UUCCUUCUGAGCCUGUGUCAACGGCAAUUUUU-GCAUUUUAAUUGCC--UGACAGGCUAAUUGCUGCCGUAGUUGUGGUAGUUGUUG-----UUUCUUCUGCCAG ......((((((((..((((..(((((((...-.......)))))))--)))))))))(((.(((((((......))))))).))).-----..........))) ( -27.50, z-score = -2.13, R) >dp4.chr3 5260216 92 - 19779522 ------CUCUGACUGUGUCUCUAACAAUUUUU-GCAUUUUAAUUGCC--UGACGGGCUAAUGGCUGUCGUAGUU-UCGUAGUUGUGUUU---AUUCCUGUGUCAG ------..(((((.........((((....((-((.....((((((.--.(((((.(....).)))))))))))-..))))...)))).---........))))) ( -19.97, z-score = -1.47, R) >droPer1.super_2 5466362 92 - 9036312 ------CUCUGACUGUGUCUCUAACAAUUUUU-GCAUUUUAAUUGCC--UGACGGGCUAAUGGCUGUCGUAGUU-UUGUAGUUGUGUUU---GUUCCUGUGUCAG ------..(((((.........((((....((-(((....((((((.--.(((((.(....).)))))))))))-.)))))...)))).---........))))) ( -22.47, z-score = -2.35, R) >droWil1.scaffold_180699 1328348 97 - 2593675 UUCCUCUUCGCCUUCUGUCAACAGCAAUUUUU-GGAUUUUAAUUGCC--UGACAGGCUGAUUGCUACCUCCACU-UAGUUCCUGCCUUU---UGCCAUGGCCCA- .........(((...(((((...((((((...-.......)))))).--)))))(((.....((((........-))))....)))...---......)))...- ( -19.30, z-score = -0.84, R) >droGri2.scaffold_15112 4638776 88 + 5172618 ---------------UGUCAGCGGUAAUUUUUAGCAUUUUAAUUGCCCUUGGCAGGCUAAUUGCGAAGACU-UUGCUGAACAUGCUUCAAGACUUCAAGACUUA- ---------------((((((.(((((((...........))))))).)))))).((.....))((((.((-(.((.......))...))).))))........- ( -19.80, z-score = -0.30, R) >consensus UUCCUUCUCGGCCUGUGUCAACGGCAAUUUUU_GCAUUUUAAUUGCC__UGACAGGCUAAUUGCUGCCGUAGUUGUGGUAGUUGCUGUU___UUUCUUCUGCCAA ...............(((((...((((((...........))))))...)))))(((.....(((((((......))))))).)))................... (-11.56 = -12.46 + 0.89)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:23:10 2011