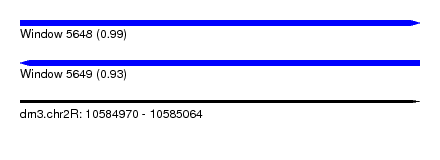
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,584,970 – 10,585,064 |
| Length | 94 |
| Max. P | 0.993733 |

| Location | 10,584,970 – 10,585,064 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 76.15 |
| Shannon entropy | 0.49919 |
| G+C content | 0.43003 |
| Mean single sequence MFE | -27.75 |
| Consensus MFE | -15.00 |
| Energy contribution | -15.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.993733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
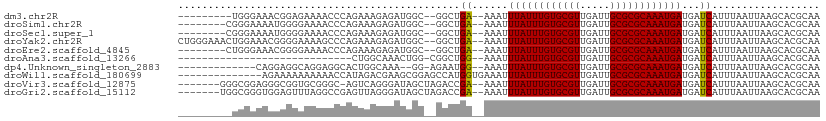
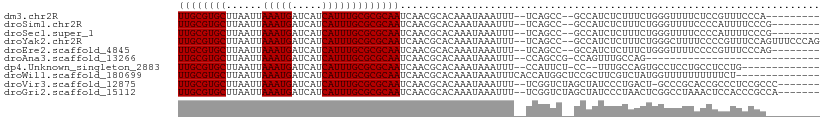
>dm3.chr2R 10584970 94 + 21146708 ---------UGGGAAACGGAGAAAACCCAGAAAGAGAUGGC--GGCUGA--AAAUUUAUUUGUGCGUUGAUUGCGCGCAAAUGAUGAUCAUUUAAUUAAGCACGCAA ---------((((............))))..........((--((((..--....((((((((((((.....))))))))))))((((......))))))).))).. ( -25.60, z-score = -2.16, R) >droSim1.chr2R 9349142 95 + 19596830 --------CGGGAAAAUGGGGAAAACCCAGAAAGAGAUGGC--GGCUGA--AAAUUUAUUUGUGCGUUGAUUGCGCGCAAAUGAUGAUCAUUUAAUUAAGCACGCAA --------........((((.....))))..........((--((((..--....((((((((((((.....))))))))))))((((......))))))).))).. ( -28.30, z-score = -2.86, R) >droSec1.super_1 8105986 95 + 14215200 --------CGGGAAAAUGGGGAAAACCCAGAAAGAGAUGGC--GGCUGA--AAAUUUAUUUGUGCGUUGAUUGCGCGCAAAUGAUGAUCAUUUAAUUAAGCACGCAA --------........((((.....))))..........((--((((..--....((((((((((((.....))))))))))))((((......))))))).))).. ( -28.30, z-score = -2.86, R) >droYak2.chr2R 10517626 103 + 21139217 CUGGGAAACUGGAAACGGGGAAAAGCCCAGAAAGAGAUGGC--GGCUGA--AAAUUUAUUUGUGCGUUGAUUGCGCGCAAAUGAUGAUCAUUUAAUUAAGCACGCAA (((((...(((....))).......))))).........((--((((..--....((((((((((((.....))))))))))))((((......))))))).))).. ( -35.10, z-score = -4.24, R) >droEre2.scaffold_4845 7325751 95 - 22589142 --------CUGGGAAACGGGGAAAACCCAGAAAGAGAUGGC--GGCUGA--AAAUUUAUUUGUGCGUUGAUUGCGCGCAAAUGAUGAUCAUUUAAUUAAGCACGCAA --------...(....)(((.....)))...........((--((((..--....((((((((((((.....))))))))))))((((......))))))).))).. ( -29.10, z-score = -3.07, R) >droAna3.scaffold_13266 5067506 75 - 19884421 -----------------------------CUGGCAAACUGG-CGGCUGG--AAAUUUAUUUGUGCGUUGAUUGCGCGCAAAUGAUGAUCAUUUAAUUAAGCACGCAA -----------------------------...........(-(((((.(--(...((((((((((((.....))))))))))))...)).........))).))).. ( -22.90, z-score = -1.79, R) >dp4.Unknown_singleton_2883 656 89 - 852 -------------CAGGAGGCAGGAGGCACUGGCAAA--GG-AGAAUGG--AAAUUUAUUUGUGCGUUGAUUGCGCGCAAAUGAUGAUCAUUUAAUUAAGCACGCAA -------------(((...((.....)).)))((..(--(.-.((((((--....((((((((((((.....))))))))))))...))))))..))..))...... ( -24.60, z-score = -2.28, R) >droWil1.scaffold_180699 1300659 93 + 2593675 --------------AGAAAAAAAAAACCAUAGACGAAGCGGAGCCAUGGUGAAAUUUAUUUGUGCGUUGAUUGCGCGCAAAUGAUGAUCAUUUAAUUAAGCACGCAA --------------.......................(((..((...(((((...((((((((((((.....))))))))))))...))))).......)).))).. ( -24.20, z-score = -2.15, R) >droVir3.scaffold_12875 14994474 97 - 20611582 -------GGGCGGAGGGCGGUGCGGGC-AGUCAGGGAUAGCUAGACCGA--AAAUUUAUUUGUGCGUUGAUUGCGCGCAAAUGAUGAUCAUUUAAUUAAGCACGCAA -------..(((....(((((.(.(((-.(((...))).))).))))((--....((((((((((((.....))))))))))))...))..........)).))).. ( -30.30, z-score = -1.62, R) >droGri2.scaffold_15112 4606204 98 - 5172618 -------UGGCGGGUGGAGUUUAGGCCGAGUUAGGGAUAGCUAGACCGA--AAAUUUAUUUGUGCGUUGAUUGCGCGCAAAUGAUGAUCAUUUAAUUAAGCACGCAA -------..((((((..((((....((......))...))))..)))((--....((((((((((((.....))))))))))))...)).............))).. ( -29.10, z-score = -1.94, R) >consensus _________GGGGAAAGGGGGAAAACCCAGAAAGAGAUGGC__GGCUGA__AAAUUUAUUUGUGCGUUGAUUGCGCGCAAAUGAUGAUCAUUUAAUUAAGCACGCAA .......................................................((((((((((((.....))))))))))))....................... (-15.00 = -15.00 + 0.00)
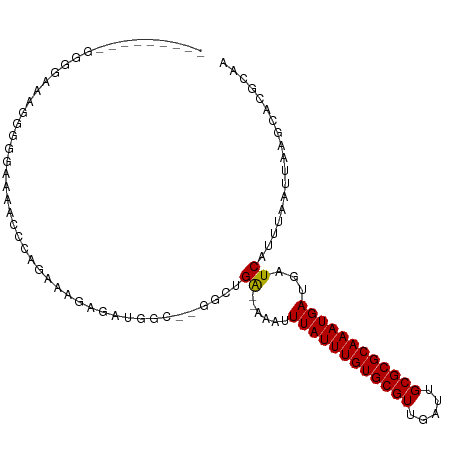
| Location | 10,584,970 – 10,585,064 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 76.15 |
| Shannon entropy | 0.49919 |
| G+C content | 0.43003 |
| Mean single sequence MFE | -16.99 |
| Consensus MFE | -12.80 |
| Energy contribution | -12.80 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.927489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
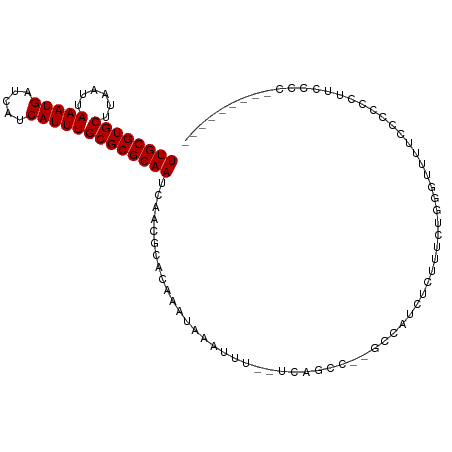
>dm3.chr2R 10584970 94 - 21146708 UUGCGUGCUUAAUUAAAUGAUCAUCAUUUGCGCGCAAUCAACGCACAAAUAAAUUU--UCAGCC--GCCAUCUCUUUCUGGGUUUUCUCCGUUUCCCA--------- ((((((((......(((((.....)))))))))))))...................--......--.............(((............))).--------- ( -15.90, z-score = -0.61, R) >droSim1.chr2R 9349142 95 - 19596830 UUGCGUGCUUAAUUAAAUGAUCAUCAUUUGCGCGCAAUCAACGCACAAAUAAAUUU--UCAGCC--GCCAUCUCUUUCUGGGUUUUCCCCAUUUUCCCG-------- ((((((((......(((((.....)))))))))))))...................--......--............((((.....))))........-------- ( -17.10, z-score = -1.35, R) >droSec1.super_1 8105986 95 - 14215200 UUGCGUGCUUAAUUAAAUGAUCAUCAUUUGCGCGCAAUCAACGCACAAAUAAAUUU--UCAGCC--GCCAUCUCUUUCUGGGUUUUCCCCAUUUUCCCG-------- ((((((((......(((((.....)))))))))))))...................--......--............((((.....))))........-------- ( -17.10, z-score = -1.35, R) >droYak2.chr2R 10517626 103 - 21139217 UUGCGUGCUUAAUUAAAUGAUCAUCAUUUGCGCGCAAUCAACGCACAAAUAAAUUU--UCAGCC--GCCAUCUCUUUCUGGGCUUUUCCCCGUUUCCAGUUUCCCAG ((((((((......(((((.....)))))))))))))...................--......--...........(((((....................))))) ( -16.95, z-score = -0.53, R) >droEre2.scaffold_4845 7325751 95 + 22589142 UUGCGUGCUUAAUUAAAUGAUCAUCAUUUGCGCGCAAUCAACGCACAAAUAAAUUU--UCAGCC--GCCAUCUCUUUCUGGGUUUUCCCCGUUUCCCAG-------- ((((((((......(((((.....)))))))))))))...................--......--...........(((((............)))))-------- ( -18.50, z-score = -1.65, R) >droAna3.scaffold_13266 5067506 75 + 19884421 UUGCGUGCUUAAUUAAAUGAUCAUCAUUUGCGCGCAAUCAACGCACAAAUAAAUUU--CCAGCCG-CCAGUUUGCCAG----------------------------- ((((((((......(((((.....)))))))))))))...................--.......-............----------------------------- ( -12.80, z-score = 0.14, R) >dp4.Unknown_singleton_2883 656 89 + 852 UUGCGUGCUUAAUUAAAUGAUCAUCAUUUGCGCGCAAUCAACGCACAAAUAAAUUU--CCAUUCU-CC--UUUGCCAGUGCCUCCUGCCUCCUG------------- ((((((((......(((((.....))))))))))))).....((((..........--.......-..--.......)))).............------------- ( -15.57, z-score = -1.78, R) >droWil1.scaffold_180699 1300659 93 - 2593675 UUGCGUGCUUAAUUAAAUGAUCAUCAUUUGCGCGCAAUCAACGCACAAAUAAAUUUCACCAUGGCUCCGCUUCGUCUAUGGUUUUUUUUUUCU-------------- ((((((((......(((((.....)))))))))))))....................((((((((........).)))))))...........-------------- ( -19.90, z-score = -1.99, R) >droVir3.scaffold_12875 14994474 97 + 20611582 UUGCGUGCUUAAUUAAAUGAUCAUCAUUUGCGCGCAAUCAACGCACAAAUAAAUUU--UCGGUCUAGCUAUCCCUGACU-GCCCGCACCGCCCUCCGCCC------- ..((((((.................(((((.(((.......))).)))))......--.(((((.((......))))))-)...))).))).........------- ( -18.60, z-score = -0.90, R) >droGri2.scaffold_15112 4606204 98 + 5172618 UUGCGUGCUUAAUUAAAUGAUCAUCAUUUGCGCGCAAUCAACGCACAAAUAAAUUU--UCGGUCUAGCUAUCCCUAACUCGGCCUAAACUCCACCCGCCA------- ((((((((......(((((.....)))))))))))))...................--..((((.((.((....)).)).))))................------- ( -17.50, z-score = -1.25, R) >consensus UUGCGUGCUUAAUUAAAUGAUCAUCAUUUGCGCGCAAUCAACGCACAAAUAAAUUU__UCAGCC__GCCAUCUCUUUCUGGGUUUUCCCCCCUUCCCC_________ ((((((((......(((((.....)))))))))))))...................................................................... (-12.80 = -12.80 + -0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:23:08 2011