
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,567,816 – 10,567,915 |
| Length | 99 |
| Max. P | 0.587619 |

| Location | 10,567,816 – 10,567,915 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 77.14 |
| Shannon entropy | 0.44572 |
| G+C content | 0.51314 |
| Mean single sequence MFE | -27.37 |
| Consensus MFE | -13.59 |
| Energy contribution | -14.49 |
| Covariance contribution | 0.91 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.587619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10567816 99 + 21146708 -CGGGAGUACAGGUGCUCUUAACGCGCUGCUAAUUUCAAUUUGUGGCAAAUAUUGACACAGCCAAAGGGAGGGGGGGGGGUUUGAUGUUGCAGCAGAGGU -.(((((((....))))))).....(((((.(((.((((.(((((.((.....)).)))))((........))........)))).))))))))...... ( -24.20, z-score = 0.23, R) >droSim1.chr2R 9330885 82 + 19596830 -CGGGAGUACAGGUGCUCUUAACGCGCUGCUAAUUUCAAUUUGUGGCAAAUAUUGACACAGCCACAGGGAGG---GCAGCAGCGGU-------------- -.(((((((....)))))))....(((((((..((((...(((((((.............))))))).))))---..)))))))..-------------- ( -32.12, z-score = -3.92, R) >droSec1.super_1 8087275 82 + 14215200 -CGGGAGUACAGGUGCUCUUAACGCGCUGCUAAUUUCAAUUUGUGGCAAAUAUUGACACAGCCACAGGGGGG---GCAGCAGCGGU-------------- -.(((((((....)))))))....(((((((..(..(...(((((((.............))))))).)..)---..)))))))..-------------- ( -32.12, z-score = -3.94, R) >droYak2.chr2R 10500091 84 + 21139217 -CGGGAGUACAGGUGCUCUUAACGCGCUGCUAAUUUCAAUUUGUGGCAAAUAUUGACACAGCCACAGGGGGGCA-GCAGCAGCGGU-------------- -.(((((((....)))))))..(((((((((..(..(....((((((.............)))))).)..)..)-))))).)))..-------------- ( -33.22, z-score = -3.81, R) >droEre2.scaffold_4845 7308726 80 - 22589142 -CGGGAGUACAGGUGCUCUUAACGCGCUGCUAAUUUCAAUUUGUGGCAAAUAUUGACACAGCCACAGGGG-----GCAGCAGCGGU-------------- -.(((((((....)))))))..(((((((((........((((((((.............)))))))).)-----))))).)))..-------------- ( -30.62, z-score = -3.47, R) >droAna3.scaffold_13266 5044779 85 - 19884421 UGGGGACUACAGGUGCUCUUAACGCGCUGCUAAUUUCAAUUUGUGGCAAAUAUUGACACAGCCACAGGGGUGCA-ACAGCCGUGCU-------------- .(((.((.....)).))).....((((.(((........((((((((.............))))))))......-..))).)))).-------------- ( -23.71, z-score = 0.03, R) >droMoj3.scaffold_6496 24437118 93 + 26866924 -UAUGAGUACAGGUGCAGCUAACGCGCUGCUAAUUUCAAUUUGUGGCAAAUAUUGACACAGACACAGCACAGAAUAGGACGU---UGCGGCAGGUGU--- -.....(((....))).....((((.(((((........((((((.((.....)).))))))..((((............))---)).)))))))))--- ( -18.20, z-score = 1.80, R) >droGri2.scaffold_15112 4583837 96 - 5172618 -UGUGAGUACAGGUGCGCCUAACGCGCUGCUAAUUUCAAUUUGUGGCAAAUAUUGACACAGACACACACAAGCGAGGGACGACCAUACGCCAGGUGU--- -.((((((...((((((.....)))))))))........((((((.((.....)).)))))))))((((..(((..((....))...)))...))))--- ( -24.80, z-score = 0.01, R) >consensus _CGGGAGUACAGGUGCUCUUAACGCGCUGCUAAUUUCAAUUUGUGGCAAAUAUUGACACAGCCACAGGGGGG___GCAGCAGCGGU______________ ..((((.....(((((.......))))).....))))..((((((((.............))))))))................................ (-13.59 = -14.49 + 0.91)
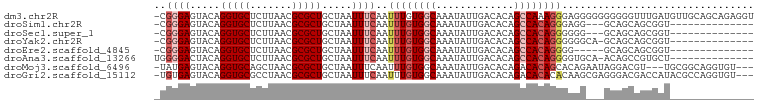

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:23:05 2011