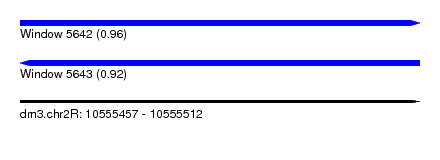
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,555,457 – 10,555,512 |
| Length | 55 |
| Max. P | 0.960201 |

| Location | 10,555,457 – 10,555,512 |
|---|---|
| Length | 55 |
| Sequences | 11 |
| Columns | 55 |
| Reading direction | forward |
| Mean pairwise identity | 95.17 |
| Shannon entropy | 0.10870 |
| G+C content | 0.29970 |
| Mean single sequence MFE | -12.07 |
| Consensus MFE | -10.98 |
| Energy contribution | -11.35 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
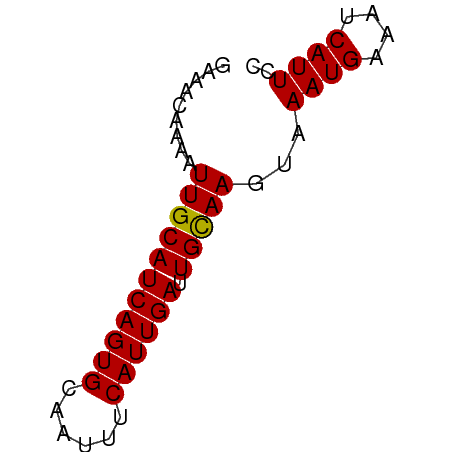
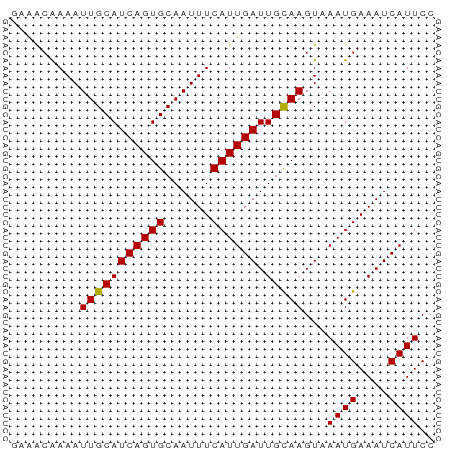
>dm3.chr2R 10555457 55 + 21146708 GGAAUGAUUUCAUUUACUUACAAUCAAUGAAAUUGCACUGAUGCAAUUUUGUUUC .(((((....)))))..........((..((((((((....))))))))..)).. ( -11.70, z-score = -2.40, R) >droSim1.chr2R 9318399 55 + 19596830 GGAAUGAUUUCAUUUACUUACAAUCAAUGAAAUUGCACUGAUGCAAUUUUGUUUC .(((((....)))))..........((..((((((((....))))))))..)).. ( -11.70, z-score = -2.40, R) >droSec1.super_1 8074983 55 + 14215200 GGAAUGAUUUCAUUUACUUACAAUCAAUGAAAUUGCACUGAUGCAAUUUUGUUUC .(((((....)))))..........((..((((((((....))))))))..)).. ( -11.70, z-score = -2.40, R) >droYak2.chr2R 10487406 55 + 21139217 GGAAUGAUUUCAUUUACUUACAAUCAAUGAAAUUGCACUGAUGCAAUUUUGUUUC .(((((....)))))..........((..((((((((....))))))))..)).. ( -11.70, z-score = -2.40, R) >droEre2.scaffold_4845 7296359 55 - 22589142 GGAAUGAUUUCAUUUACUUACAAUCAAUGAAAUUGCACUGAUGCAAUUUUGUUUC .(((((....)))))..........((..((((((((....))))))))..)).. ( -11.70, z-score = -2.40, R) >droAna3.scaffold_13266 5032697 55 - 19884421 GGAAUGAUUUCAUUUACUUGCAAUCAAUGAAAUUGCACUGAUGCAAUUUUCCCUU (((.(((((.((......)).)))))...((((((((....)))))))))))... ( -12.10, z-score = -2.43, R) >dp4.chr3 5205772 55 + 19779522 GGAAUGAUUUCAUUUACUUGCAAUCAAUGAAAUUGCACUGAUGCAAUUUUGUUUU ....(((((.((......)).)))))(..((((((((....))))))))..)... ( -12.10, z-score = -1.79, R) >droPer1.super_2 5413353 55 + 9036312 GGAAUGAUUUCAUUUACUUGCAAUCAAUGAAAUUGCACUGAUGCAAUUUUGUUUU ....(((((.((......)).)))))(..((((((((....))))))))..)... ( -12.10, z-score = -1.79, R) >droWil1.scaffold_180699 1267646 54 + 2593675 -GAAUGAUUUCAUUUACUUGCAAUCAAUGAAAUUGCACUGAUGCAAUUUUGUUUU -...(((((.((......)).)))))(..((((((((....))))))))..)... ( -12.10, z-score = -1.98, R) >droMoj3.scaffold_6496 24421249 52 + 26866924 GGAAUGAUUUCAUUUACUUGCAAUCAAUGAAAUUGCACUGAUGCAAUUUCGC--- ....(((((.((......)).)))))..(((((((((....)))))))))..--- ( -13.80, z-score = -2.74, R) >droGri2.scaffold_15112 4569521 55 - 5172618 GGAAUGAUUUCAUUUACUUGCAAUCAAUGAAAUUGCACUGAUGCAAUUUUGUUUG ....(((((.((......)).)))))(..((((((((....))))))))..)... ( -12.10, z-score = -1.90, R) >consensus GGAAUGAUUUCAUUUACUUGCAAUCAAUGAAAUUGCACUGAUGCAAUUUUGUUUC .(((((....)))))..........((((((((((((....)))))))))))).. (-10.98 = -11.35 + 0.37)

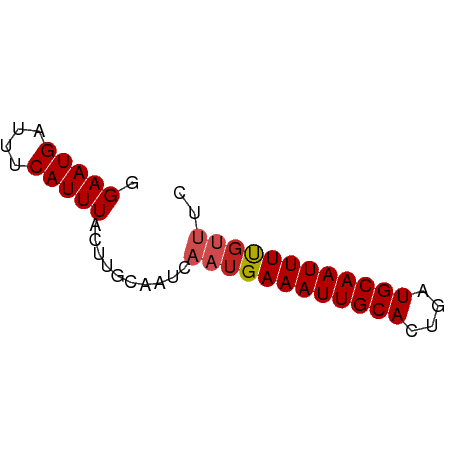
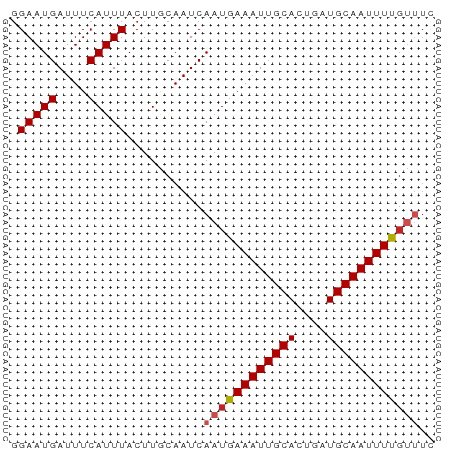
| Location | 10,555,457 – 10,555,512 |
|---|---|
| Length | 55 |
| Sequences | 11 |
| Columns | 55 |
| Reading direction | reverse |
| Mean pairwise identity | 95.17 |
| Shannon entropy | 0.10870 |
| G+C content | 0.29970 |
| Mean single sequence MFE | -10.05 |
| Consensus MFE | -9.94 |
| Energy contribution | -9.69 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.918443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
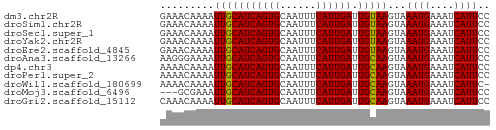
>dm3.chr2R 10555457 55 - 21146708 GAAACAAAAUUGCAUCAGUGCAAUUUCAUUGAUUGUAAGUAAAUGAAAUCAUUCC .........(((((((((((......)))))).)))))...((((....)))).. ( -8.60, z-score = -1.06, R) >droSim1.chr2R 9318399 55 - 19596830 GAAACAAAAUUGCAUCAGUGCAAUUUCAUUGAUUGUAAGUAAAUGAAAUCAUUCC .........(((((((((((......)))))).)))))...((((....)))).. ( -8.60, z-score = -1.06, R) >droSec1.super_1 8074983 55 - 14215200 GAAACAAAAUUGCAUCAGUGCAAUUUCAUUGAUUGUAAGUAAAUGAAAUCAUUCC .........(((((((((((......)))))).)))))...((((....)))).. ( -8.60, z-score = -1.06, R) >droYak2.chr2R 10487406 55 - 21139217 GAAACAAAAUUGCAUCAGUGCAAUUUCAUUGAUUGUAAGUAAAUGAAAUCAUUCC .........(((((((((((......)))))).)))))...((((....)))).. ( -8.60, z-score = -1.06, R) >droEre2.scaffold_4845 7296359 55 + 22589142 GAAACAAAAUUGCAUCAGUGCAAUUUCAUUGAUUGUAAGUAAAUGAAAUCAUUCC .........(((((((((((......)))))).)))))...((((....)))).. ( -8.60, z-score = -1.06, R) >droAna3.scaffold_13266 5032697 55 + 19884421 AAGGGAAAAUUGCAUCAGUGCAAUUUCAUUGAUUGCAAGUAAAUGAAAUCAUUCC ...((((..(((((((((((......)))))).))))).....((....)))))) ( -11.70, z-score = -1.54, R) >dp4.chr3 5205772 55 - 19779522 AAAACAAAAUUGCAUCAGUGCAAUUUCAUUGAUUGCAAGUAAAUGAAAUCAUUCC .........(((((((((((......)))))).)))))...((((....)))).. ( -10.60, z-score = -1.66, R) >droPer1.super_2 5413353 55 - 9036312 AAAACAAAAUUGCAUCAGUGCAAUUUCAUUGAUUGCAAGUAAAUGAAAUCAUUCC .........(((((((((((......)))))).)))))...((((....)))).. ( -10.60, z-score = -1.66, R) >droWil1.scaffold_180699 1267646 54 - 2593675 AAAACAAAAUUGCAUCAGUGCAAUUUCAUUGAUUGCAAGUAAAUGAAAUCAUUC- .........(((((((((((......)))))).)))))...((((....)))).- ( -10.60, z-score = -1.48, R) >droMoj3.scaffold_6496 24421249 52 - 26866924 ---GCGAAAUUGCAUCAGUGCAAUUUCAUUGAUUGCAAGUAAAUGAAAUCAUUCC ---..(((((((((....)))))))))..(((((.((......)).))))).... ( -13.50, z-score = -2.46, R) >droGri2.scaffold_15112 4569521 55 + 5172618 CAAACAAAAUUGCAUCAGUGCAAUUUCAUUGAUUGCAAGUAAAUGAAAUCAUUCC .........(((((((((((......)))))).)))))...((((....)))).. ( -10.60, z-score = -1.73, R) >consensus GAAACAAAAUUGCAUCAGUGCAAUUUCAUUGAUUGCAAGUAAAUGAAAUCAUUCC .........(((((((((((......)))))).)))))...((((....)))).. ( -9.94 = -9.69 + -0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:23:04 2011