| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,532,658 – 10,532,751 |
| Length | 93 |
| Max. P | 0.792123 |

| Location | 10,532,658 – 10,532,751 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 77.28 |
| Shannon entropy | 0.43351 |
| G+C content | 0.40497 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -12.01 |
| Energy contribution | -12.43 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.692288 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
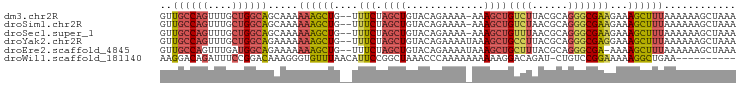
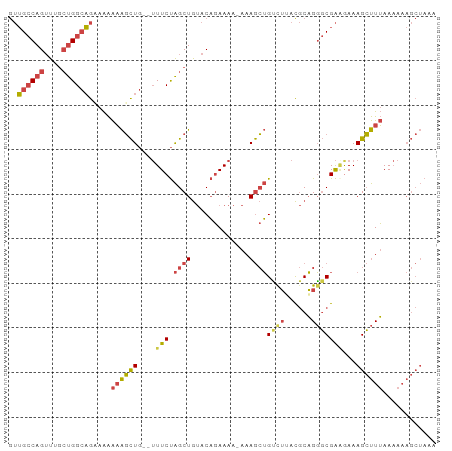
>dm3.chr2R 10532658 93 + 21146708 UUUAGCUUUUUUAAAGCUUUCUUCGCCCUGCGUAAGACAGCUUU-UUUUCUGUACAGCUAGAAA--CAGCUUUUUUGCUGCCAGCAAACUGGCAAC (((((((.....((((((.(((((((...))).)))).))))))-..........)))))))..--.(((......)))(((((....)))))... ( -27.46, z-score = -2.33, R) >droSim1.chr2R 9295786 93 + 19596830 UUUAGCUUUUUUAAAGCUUUCUUCGCCCUGCGUUAGACAGCUUU-UUUUCUGUACAGCUAGAAA--CAGCUUUUUUGCUGCCAGCAAACUGGCAAC (((((((.....((((((.(((.(((...)))..))).))))))-..........)))))))..--.(((......)))(((((....)))))... ( -25.96, z-score = -1.92, R) >droSec1.super_1 8052249 93 + 14215200 UUUAGCUUUUUUAAAGCUUUCUUCGCCCUGCGUUAAACAGCUUU-UUUUCUGUACAGCUAGAAA--CAGCUUUUUUGCUGCCAGCAAACUGGCAAC (((((((.....((((((.....(((...)))......))))))-..........)))))))..--.(((......)))(((((....)))))... ( -22.06, z-score = -0.88, R) >droYak2.chr2R 10463844 94 + 21139217 UUUAGCUUUUUUAAAGCUUUCCUCGCCCUGCGUAAGGCAGCUUUAUUUUCUGUACAGCUAGAAA--CAGCUUUUUUUCUGCCAGCAAACUGGCAAC ...((((.((((...(((((...(((...))).)))))((((.(((.....))).)))).))))--.)))).......((((((....)))))).. ( -24.20, z-score = -1.30, R) >droEre2.scaffold_4845 7270648 93 - 22589142 UUUAGCUUUUUUAAAGCUUUU-UCGCCCUGCGUAAAGCAGCUUUAUUUUCUGUACAGCUAGAAA--CAGCUUUUUUUCUGCCAUCAAACUGGCAAC ...((((.((((...(((((.-.(((...))).)))))((((.(((.....))).)))).))))--.)))).......(((((......))))).. ( -22.50, z-score = -1.94, R) >droWil1.scaffold_181140 1059241 85 + 1556085 ----------UUCAGCCUUUUUCCGGACAG-AUCUGUCCUUUUUUUUUUGGGUUUAGCCGGAAUGUUAAACACCCUUUGUCCGGAAAUCUGUCCUU ----------..(((....(((((((((((-(.....((..........))(((((((......)))))))....)))))))))))).)))..... ( -24.70, z-score = -2.73, R) >consensus UUUAGCUUUUUUAAAGCUUUCUUCGCCCUGCGUAAGACAGCUUU_UUUUCUGUACAGCUAGAAA__CAGCUUUUUUGCUGCCAGCAAACUGGCAAC ...(((((.....))))).............(..(((.((((....((((((......))))))...)))).)))..)((((((....)))))).. (-12.01 = -12.43 + 0.42)



| Location | 10,532,658 – 10,532,751 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 77.28 |
| Shannon entropy | 0.43351 |
| G+C content | 0.40497 |
| Mean single sequence MFE | -27.32 |
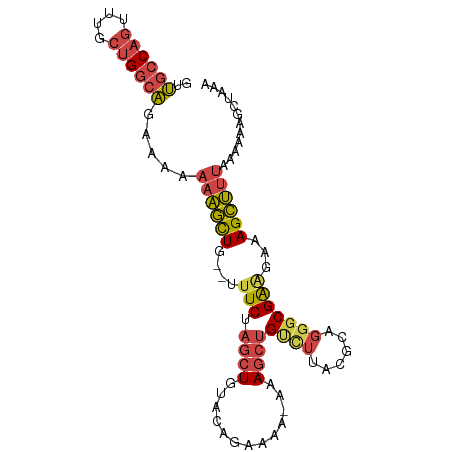
| Consensus MFE | -16.43 |
| Energy contribution | -17.10 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.792123 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 10532658 93 - 21146708 GUUGCCAGUUUGCUGGCAGCAAAAAAGCUG--UUUCUAGCUGUACAGAAAA-AAAGCUGUCUUACGCAGGGCGAAGAAAGCUUUAAAAAAGCUAAA ((((((((....)))))))).....((((.--(((((........))))).-((((((.((((.(((...))))))).)))))).....))))... ( -32.20, z-score = -2.63, R) >droSim1.chr2R 9295786 93 - 19596830 GUUGCCAGUUUGCUGGCAGCAAAAAAGCUG--UUUCUAGCUGUACAGAAAA-AAAGCUGUCUAACGCAGGGCGAAGAAAGCUUUAAAAAAGCUAAA ((((((((....)))))))).....((((.--(((((........))))).-((((((.(((..(((...))).))).)))))).....))))... ( -29.50, z-score = -1.88, R) >droSec1.super_1 8052249 93 - 14215200 GUUGCCAGUUUGCUGGCAGCAAAAAAGCUG--UUUCUAGCUGUACAGAAAA-AAAGCUGUUUAACGCAGGGCGAAGAAAGCUUUAAAAAAGCUAAA ((((((((....)))))))).....((((.--(((((........))))).-((((((......(((...))).....)))))).....))))... ( -27.20, z-score = -1.09, R) >droYak2.chr2R 10463844 94 - 21139217 GUUGCCAGUUUGCUGGCAGAAAAAAAGCUG--UUUCUAGCUGUACAGAAAAUAAAGCUGCCUUACGCAGGGCGAGGAAAGCUUUAAAAAAGCUAAA .(((((((....)))))))......((((.--(((((........))))).(((((((..(((.(((...))))))..)))))))....))))... ( -28.10, z-score = -1.15, R) >droEre2.scaffold_4845 7270648 93 + 22589142 GUUGCCAGUUUGAUGGCAGAAAAAAAGCUG--UUUCUAGCUGUACAGAAAAUAAAGCUGCUUUACGCAGGGCGA-AAAAGCUUUAAAAAAGCUAAA .(((((..((((((((((((((........--))))).))))).))))........((((.....)))))))))-...(((((.....)))))... ( -24.60, z-score = -1.07, R) >droWil1.scaffold_181140 1059241 85 - 1556085 AAGGACAGAUUUCCGGACAAAGGGUGUUUAACAUUCCGGCUAAACCCAAAAAAAAAAGGACAGAU-CUGUCCGGAAAAAGGCUGAA---------- .....(((.(((((((((...((((......(.....).....))))..........(((....)-)))))))))))....)))..---------- ( -22.30, z-score = -1.49, R) >consensus GUUGCCAGUUUGCUGGCAGAAAAAAAGCUG__UUUCUAGCUGUACAGAAAA_AAAGCUGUCUUACGCAGGGCGAAGAAAGCUUUAAAAAAGCUAAA ..((((((....)))))).....((((((...((((.((((.............))))((((......))))))))..))))))............ (-16.43 = -17.10 + 0.67)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:23:01 2011