
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,513,346 – 10,513,449 |
| Length | 103 |
| Max. P | 0.542742 |

| Location | 10,513,346 – 10,513,449 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 77.77 |
| Shannon entropy | 0.43977 |
| G+C content | 0.35879 |
| Mean single sequence MFE | -22.55 |
| Consensus MFE | -11.80 |
| Energy contribution | -11.57 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.542742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
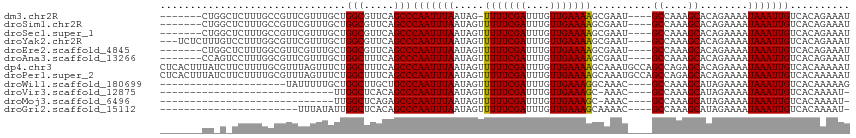
>dm3.chr2R 10513346 103 - 21146708 -------CUGGCUCUUUGCCGUUCGUUUGCUGGCGUUCAGCCCAAUUUAAUAG-UUUUCGAUUUGUUGAAAAGCGAAU----GCCAAAGCACAGAAAAUAAAUUGUCACAGAAAU -------(((...(((((.(((((((((...(((.....)))...((((((((-........))))))))))))))))----).)))))..)))..................... ( -25.40, z-score = -1.24, R) >droSim1.chr2R 9275616 104 - 19596830 -------CUGGCUCUUUGCCGUUCGUUUGCUGGCGUUCAGCCCAAUUUAAUAGUUUUUCGAUUUGUUGAAAAGCGAAU----GCCAAAGCACAGAAAAUAAAUUGUCACAGAAAU -------(((...(((((.(((((.......(((.....)))..........(((((((((....)))))))))))))----).)))))..)))..................... ( -25.10, z-score = -1.08, R) >droSec1.super_1 8030398 104 - 14215200 -------CUGGCUCUUUGCCGUUCGUUUGCUGGCGUUCAGCCCAAUUUAAUAGUUUUUCGAUUUGUUGAAAAGCGAAU----GCCAAAGCACAGAAAAUAAAUUGUCACAGAAAU -------(((...(((((.(((((.......(((.....)))..........(((((((((....)))))))))))))----).)))))..)))..................... ( -25.10, z-score = -1.08, R) >droYak2.chr2R 10442077 108 - 21139217 ---UCUCUUUGUCCUUUGGCGUUCGUUUGCUGGCGUUCAGCCCAAUUUAAUAGUUUUUCGAUUUGUUGAAAAGCGAAU----GCCAAAGCACAGAAAAUAAAUUGUCACAGAAAU ---....(((((.(((((((((((.......(((.....)))..........(((((((((....)))))))))))))----))))))).))))).................... ( -33.60, z-score = -3.98, R) >droEre2.scaffold_4845 7249378 104 + 22589142 -------CUGGCUCUUUGGCGUUCGUUUGCUGGCGUUCAGCCCAAUUUAAUAGUUUUUCGAUUUGUUGAAAAGCGAAU----GCCAAAGCACAGAAAAUAAAUUGUCACAGAAAU -------(((...(((((((((((.......(((.....)))..........(((((((((....)))))))))))))----)))))))..)))..................... ( -32.20, z-score = -3.02, R) >droAna3.scaffold_13266 4992390 104 + 19884421 -------CCAGUCCUUUGGCGUUCGUUUGCUGGCUUUCAGCCCAAUUUAAUAGUUUUUCGAUUUGUUGAAAAGCGAAU----GCCAAAGCACAGAAAAUAAAUUGUCACAGAAAU -------...((.(((((((((((....((((.....))))...........(((((((((....)))))))))))))----))))))).))....................... ( -31.00, z-score = -3.66, R) >dp4.chr3 5159590 115 - 19779522 CUCACUUUAUCUUCUUUUGCGUUUAGUUUCUGGCUUUCAGCCCAAUUUAAUAGUUUUUCGAUUUGUUGAAAAGCAAAUGCCAGCCAGAGCACAGAAAAUAAAUUGUCACAAAAAU ..((.(((((.((((..(((........(((((((..((.............(((((((((....)))))))))...))..)))))))))).)))).))))).)).......... ( -21.49, z-score = -0.83, R) >droPer1.super_2 5360294 115 - 9036312 CUCACUUUAUCUUCUUUUGCGUUUAGUUUCUGGCUUUCAGCCCAAUUUAAUAGUUUUUCGAUUUGUUGAAAAGCAAAUGCCAGCCAGAGCACAGAAAAUAAAUUGUCACAAAAAU ..((.(((((.((((..(((........(((((((..((.............(((((((((....)))))))))...))..)))))))))).)))).))))).)).......... ( -21.49, z-score = -0.83, R) >droWil1.scaffold_180699 1204866 90 - 2593675 ---------------------UAUUUUUGCUGGCUUGCUGCCCAAUUUAAUAGUUUUUCGAUUUGUUGAAAGGCAAAC----GCCAAAGCAUAGAAAAUAAAUUGUCACAAAAAG ---------------------..((((((..(((.....)))(((((((...(((((((((....)))))))))....----((....))........)))))))...)))))). ( -18.80, z-score = -1.02, R) >droVir3.scaffold_12875 14900264 80 + 20611582 -----------------------------UUGGCUCACAGCCCAAUUUAAUAGUUUUUCGAUUUGUUGAAAGC-AAAC----GCCAAAGCAUAGAAAAUAAAUUGUCACAAAAU- -----------------------------..(((.....)))(((((((.....(((((....((((....((-....----))...))))..)))))))))))).........- ( -12.10, z-score = -0.69, R) >droMoj3.scaffold_6496 24355791 80 - 26866924 -----------------------------UUGGCUCAGAGCCCAAUUUAAUAGUUUUUCGAUUUGUUGAAAGC-AAAC----GCCAAAGCAUAGAAAAUAAAUUGUCACAAAAU- -----------------------------..(((.....)))(((((((.....(((((....((((....((-....----))...))))..)))))))))))).........- ( -12.10, z-score = -0.57, R) >droGri2.scaffold_15112 4523205 87 + 5172618 -----------------------UUUAUAUUGGCUCACAGCCCAAUUUAAUAGUUUUUCGAUUUGUUGAAAGCAAAAC----GCCAAAGCAUAGAAAAUAAAUUGUCACAAAAU- -----------------------........(((.....)))(((((((.....((((((.(((((.....))))).)----((....))...)))))))))))).........- ( -12.20, z-score = -0.38, R) >consensus _______CU_G_UCUUUGGCGUUCGUUUGCUGGCUUUCAGCCCAAUUUAAUAGUUUUUCGAUUUGUUGAAAAGCAAAU____GCCAAAGCACAGAAAAUAAAUUGUCACAAAAAU ...............................(((.....)))(((((((.....(((((((....)))))))..........((....))........))))))).......... (-11.80 = -11.57 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:22:59 2011