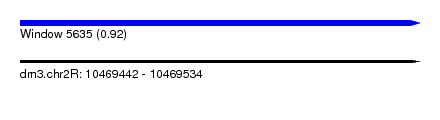
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,469,442 – 10,469,534 |
| Length | 92 |
| Max. P | 0.919618 |

| Location | 10,469,442 – 10,469,534 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 62.08 |
| Shannon entropy | 0.66065 |
| G+C content | 0.46545 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -9.42 |
| Energy contribution | -9.25 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.919618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
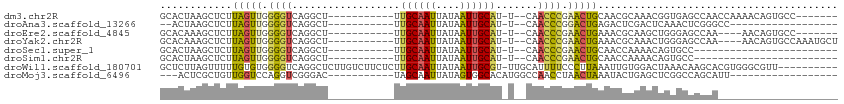
>dm3.chr2R 10469442 92 + 21146708 GCACUAAGCUCUUAGUUGGGGUCAGGCU-----------UUGCAAUUAUAAUUGCAU-U--CAACCCGAACUGCAACGCAAACGGUGAGCCAACCAAAACAGUGCC------- (((((..((...(((((.((((...(..-----------.((((((....)))))).-.--).)))).)))))....))....(((......))).....))))).------- ( -27.60, z-score = -1.90, R) >droAna3.scaffold_13266 10863078 79 + 19884421 --ACUAAGCUCUUAGUUGGGGUCAGGCU-----------UUGCAAUUAUAAUUGCAU-U--CAACCCGGACUGAGACUCGACUCAAACUCGGGCC------------------ --.......((((((((.((((...(..-----------.((((((....)))))).-.--).)))).))))))))(((((.......)))))..------------------ ( -25.60, z-score = -2.91, R) >droEre2.scaffold_4845 7204668 88 - 22589142 GCACAAAGCUCUUAGUUGGGGUCAGGCU-----------UUGCAAUUAUAAUUGCAU-U--CAACCCGAACUGAAACGCAAGCUGGGAGCCAA----AACAGUGCC------- ((((...((((((((((((..((((...-----------.((((((....))))))(-(--(.....)))))))..).).))))))))))...----....)))).------- ( -27.50, z-score = -1.81, R) >droYak2.chr2R 10398089 95 + 21139217 GCACAAAGCUCUUAGUUGGGGUCAGGCU-----------UUGCAAUUAUAAUUGCAU-U--CAACCCGAACUGAAACGCAAACUGGGAGCCAA----AACAGUGCCAAAUGCU ((((...((((((((((((..((((...-----------.((((((....))))))(-(--(.....)))))))..).).))))))))))...----....))))........ ( -27.50, z-score = -1.57, R) >droSec1.super_1 7986628 75 + 14215200 GCACUAAGCUCUUAGUUGGGGUCAGGCU-----------UUGCAAUUAUAAUUGCAU-U--CAACCCGAACUGCAACCAAAACAGUGCC------------------------ (((((.......(((((.((((...(..-----------.((((((....)))))).-.--).)))).)))))..........))))).------------------------ ( -20.33, z-score = -1.59, R) >droSim1.chr2R 9230906 75 + 19596830 GCACUAAGCUCUUAGUUGGGGUCAGGCU-----------UUGCAAUUAUAAUUGCAU-U--CAACCCGAACUGCAACCAAAACAGUGCC------------------------ (((((.......(((((.((((...(..-----------.((((((....)))))).-.--).)))).)))))..........))))).------------------------ ( -20.33, z-score = -1.59, R) >droWil1.scaffold_180701 1046585 102 + 3904529 GCUCUUAGUUUUUGUGUGGGGUCAGGCUCUUGUCUUCUCUUGCAAUUAUAAUUGCGU-UUGCAUUUUCCCUUAAAUUGUGGACUAAACAAGCACGUGGGCGUU---------- ((((...((.((((((..(((..((((....))))..)))..)............((-(..((.(((.....))).))..)))...))))).))..))))...---------- ( -25.60, z-score = -1.24, R) >droMoj3.scaffold_6496 17002122 81 + 26866924 ---ACUCGCUGUUGGUCCAGGUCGGGAC-----------UAGCAAUUAUAGUGGCACAUGGCCAACCUAACUAAAUACUGAGCUCGGCCAGCAUU------------------ ---......((((((((.((.((((.((-----------((.......))))(((.....)))..............)))).)).))))))))..------------------ ( -25.50, z-score = -1.40, R) >consensus GCACUAAGCUCUUAGUUGGGGUCAGGCU___________UUGCAAUUAUAAUUGCAU_U__CAACCCGAACUGAAACGCAAACUGGGCCCCAA____________________ ............(((((.((((..................((((((....)))))).......)))).)))))........................................ ( -9.42 = -9.25 + -0.17)
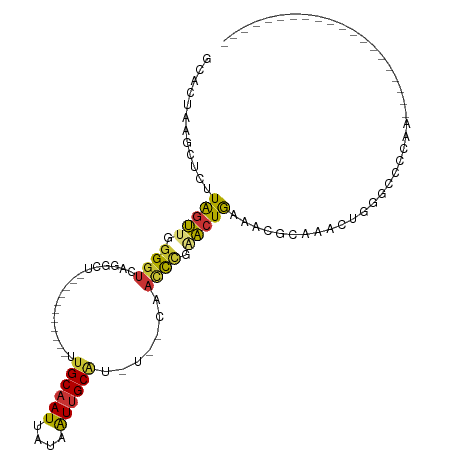
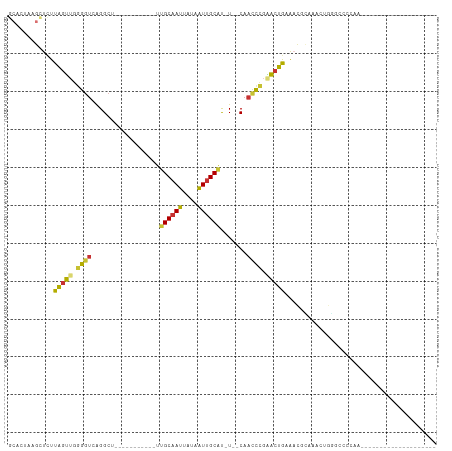
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:22:57 2011