| Sequence ID | dm3.chr2R |
|---|---|
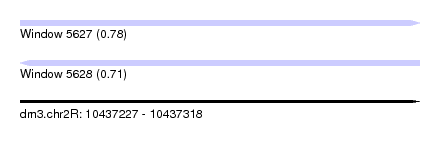
| Location | 10,437,227 – 10,437,318 |
| Length | 91 |
| Max. P | 0.781096 |

| Location | 10,437,227 – 10,437,318 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 72.18 |
| Shannon entropy | 0.50041 |
| G+C content | 0.47707 |
| Mean single sequence MFE | -18.52 |
| Consensus MFE | -7.99 |
| Energy contribution | -8.77 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.781096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
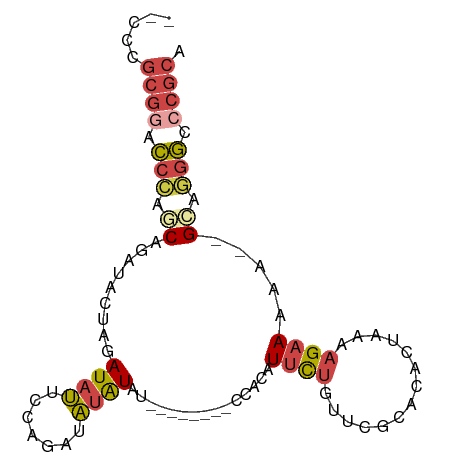
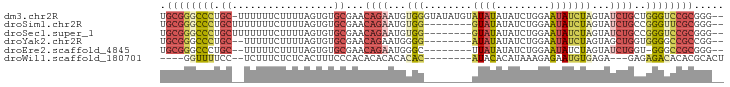
>dm3.chr2R 10437227 91 + 21146708 --CCCGCGGACCCAGCAGAUACUAGAUAUUCCAGAUAUAUAUACAUAUACCCACAUUCUGUUCGCACACUAAAAGAAAAAA-GCAGGGCCCGCA --...((((.(((((((((..((.(.....).)).(((((....))))).......)))))).((................-)).))).)))). ( -18.69, z-score = -2.49, R) >droSim1.chr2R 9194938 84 + 19596830 --CCCGCGAACCCGGCAGAUACUAGAUAUUCCAGAUAUAUAC--------CCACAUUCUGUUCGCACACUAAAAGAAAAAAAGCAGGGCCCGCA --...(((..(((((((((......((((.......))))..--------......)))))).((.................)).)))..))). ( -15.07, z-score = -1.06, R) >droSec1.super_1 7951126 84 + 14215200 --CCCGCGGACCCGGCAGAUACUAGAUAUUCCAGAUAUAUAC--------CCACAUUCUGUUCGCACACUAAAAGAAAAAAAGCAGGGCCCGCA --...((((.(((((((((......((((.......))))..--------......)))))).((.................)).))).)))). ( -18.87, z-score = -1.76, R) >droYak2.chr2R 10365595 82 + 21139217 --CCGGCGGCCCCACCAGCUACUAGAUAUUCCAGAUAUAUAU--------CCCCAUUCUGUUCGCACACUAAAAGAAAAA--GCAGGGCCCGCA --...((((.(((..(((......(((((.........))))--------)......)))...((...............--)).))).)))). ( -16.96, z-score = -0.62, R) >droEre2.scaffold_4845 7172456 81 - 22589142 --CCCGCGGCCC-ACCAGAUACUAGAUAUUCCAGAUAUAUAA--------GCCCAUUCUGUUCGCACACUAAAAGAAAAA--GCAGGGCCCGCA --...(((((((-..((((......((((.......))))..--------......))))...((...............--)).))).)))). ( -17.00, z-score = -1.45, R) >droWil1.scaffold_180701 1006299 77 + 3904529 AGUGCGUGUGUCUCUC---UCUCACAUUCUCUUUAUGUGUAU--------GUGUGUGUGUGUGGGAAAGUGAGAGAAAGA--GGAAAACC---- .....((...((((((---((((((.(((((..((((..((.--------...))..)))).))))).))))))))..))--))...)).---- ( -24.50, z-score = -4.10, R) >consensus __CCCGCGGACCCAGCAGAUACUAGAUAUUCCAGAUAUAUAU________CCACAUUCUGUUCGCACACUAAAAGAAAAA__GCAGGGCCCGCA .....((((.(((.((.........((((.......))))...............((((..............)))).....)).))).)))). ( -7.99 = -8.77 + 0.78)

| Location | 10,437,227 – 10,437,318 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 72.18 |
| Shannon entropy | 0.50041 |
| G+C content | 0.47707 |
| Mean single sequence MFE | -24.77 |
| Consensus MFE | -11.86 |
| Energy contribution | -12.62 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.712528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10437227 91 - 21146708 UGCGGGCCCUGC-UUUUUUCUUUUAGUGUGCGAACAGAAUGUGGGUAUAUGUAUAUAUAUCUGGAAUAUCUAGUAUCUGCUGGGUCCGCGGG-- .((((((((.((-............((((((..((.....))..))))))..........((((.....)))).....)).))))))))...-- ( -28.90, z-score = -1.98, R) >droSim1.chr2R 9194938 84 - 19596830 UGCGGGCCCUGCUUUUUUUCUUUUAGUGUGCGAACAGAAUGUGG--------GUAUAUAUCUGGAAUAUCUAGUAUCUGCCGGGUUCGCGGG-- .((((((((.((.............((((((..((.....))..--------))))))..((((.....)))).....)).))))))))...-- ( -26.10, z-score = -1.59, R) >droSec1.super_1 7951126 84 - 14215200 UGCGGGCCCUGCUUUUUUUCUUUUAGUGUGCGAACAGAAUGUGG--------GUAUAUAUCUGGAAUAUCUAGUAUCUGCCGGGUCCGCGGG-- .((((((((.((.............((((((..((.....))..--------))))))..((((.....)))).....)).))))))))...-- ( -28.90, z-score = -2.16, R) >droYak2.chr2R 10365595 82 - 21139217 UGCGGGCCCUGC--UUUUUCUUUUAGUGUGCGAACAGAAUGGGG--------AUAUAUAUCUGGAAUAUCUAGUAGCUGGUGGGGCCGCCGG-- .((((.(((..(--(....((.(((((((.....(((((((...--------...))).))))..))).)))).))..))..)))))))...-- ( -23.90, z-score = 0.12, R) >droEre2.scaffold_4845 7172456 81 + 22589142 UGCGGGCCCUGC--UUUUUCUUUUAGUGUGCGAACAGAAUGGGC--------UUAUAUAUCUGGAAUAUCUAGUAUCUGGU-GGGCCGCGGG-- .((((.(((.((--(..((((.((.(....).)).))))..)))--------(((.(((.((((.....))))))).))).-)))))))...-- ( -22.20, z-score = 0.05, R) >droWil1.scaffold_180701 1006299 77 - 3904529 ----GGUUUUCC--UCUUUCUCUCACUUUCCCACACACACACAC--------AUACACAUAAAGAGAAUGUGAGA---GAGAGACACACGCACU ----((....))--((((((((((((.(((.(............--------...........).))).))))))---)))))).......... ( -18.60, z-score = -3.95, R) >consensus UGCGGGCCCUGC__UUUUUCUUUUAGUGUGCGAACAGAAUGUGG________AUAUAUAUCUGGAAUAUCUAGUAUCUGCUGGGUCCGCGGG__ .((((((((.................(((....)))........................((((.....))))........))))))))..... (-11.86 = -12.62 + 0.75)
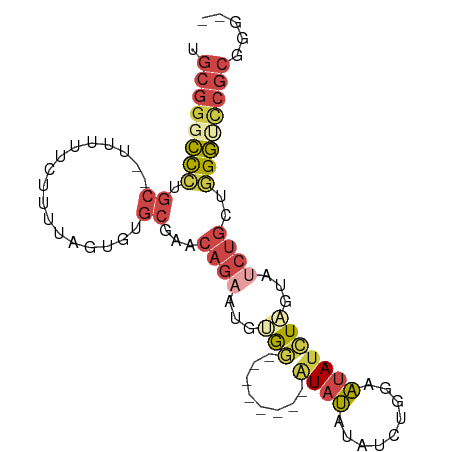
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:22:51 2011