| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,858,063 – 2,858,160 |
| Length | 97 |
| Max. P | 0.998259 |

| Location | 2,858,063 – 2,858,160 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 60.36 |
| Shannon entropy | 0.70643 |
| G+C content | 0.59096 |
| Mean single sequence MFE | -23.89 |
| Consensus MFE | -8.70 |
| Energy contribution | -7.99 |
| Covariance contribution | -0.71 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.30 |
| SVM RNA-class probability | 0.998259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
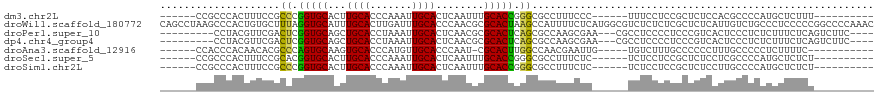
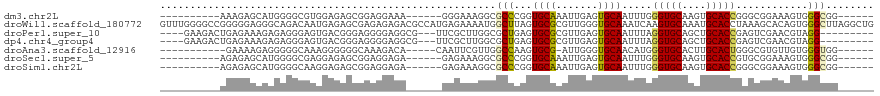
>dm3.chr2L 2858063 97 + 23011544 ------CCGCCCACUUUCCGCCCGGUGCACUUGCACCCAAAUUGCACUCAAUUUGCACCGGGCGCCUUUCCC------UUUCCUCCGCUCUCCACGCCCCAUGCUCUUU---------- ------..((........(((((((((((..((((.......)))).......)))))))))))........------........((.......)).....)).....---------- ( -25.30, z-score = -4.03, R) >droWil1.scaffold_180772 2961057 119 - 8906247 CAGCCUAAGCCCACUGUGCUUUAGGUGCAUUUGCACUUGAUUUGCACCCAACGCGCACUAAGCCAUUUUCUCAUGGCGUCUCUCUCGCUCUCAUUGUCUGCCCUCCCCCGGCCCCAAAC ..(((..........((((.((((((((....))))))))...)))).....(((......(((((......)))))........))).....................)))....... ( -29.14, z-score = -1.76, R) >droPer1.super_10 34012 103 - 3432795 ---------CCUACGUUCGACUCGGUGCAGCUGCACCUAAAUUGCACUCAACGCGCACUCAGCGCCAAGCGAA---CGCCUCCCCUCCCGUCACUCCCUCUCUUUCUCAGUCUUC---- ---------....((((((.((.(((((....)))))...............((((.....))))..))))))---)).....................................---- ( -23.10, z-score = -2.41, R) >dp4.chr4_group4 2228882 103 - 6586962 ---------CCUACGUUCGACUCGGUGCAGCUGCACCUAAAUUGCACUCAACGCGCACUCAGCGCCAAGCGAA---CGCCUCCCCUCCCGUCACUCCCUCUCUUUCUCAGUCUUC---- ---------....((((((.((.(((((....)))))...............((((.....))))..))))))---)).....................................---- ( -23.10, z-score = -2.41, R) >droAna3.scaffold_12916 722891 96 + 16180835 ------CCACCCACAACACGCCCAGUGCAAGUGCACCCAUGUUGCACCCAAU-CGCACUUGGCCAACGAAUUG-----UGUCUUUGCCCCCCUUUGCCCCCUCUUUUC----------- ------.....(((((...(((.(((((..(((((.......))))).....-.))))).))).......)))-----))............................----------- ( -18.20, z-score = -1.18, R) >droSec1.super_5 1017125 97 + 5866729 ------CCGCCCACUUUCCGCACGGUGCACUUGCACCCAAAUUGCACUCAAUUUGCACCGGGCGCCUUUCUC------UCUCCUCCGCUCUCCUCGCCCCAUGCUCUCU---------- ------.(((((...........(((((....)))))(((((((....)))))))....)))))........------.................((.....)).....---------- ( -22.50, z-score = -3.12, R) >droSim1.chr2L 2819058 97 + 22036055 ------CCGCCCACUUUCCGCCCGGUGCACUUGCACCCAAAUUGCACUCAAUUUGCACCGGGCGCCUUUCUC------UCUCCUCCGCUCUCCUUGCCCCAUGCUCUCU---------- ------..((........(((((((((((..((((.......)))).......)))))))))))........------........((.......)).....)).....---------- ( -25.90, z-score = -4.76, R) >consensus ______CCGCCCACUUUCCGCCCGGUGCACUUGCACCCAAAUUGCACUCAAUGCGCACUGGGCGCCUUUCUC______UCUCCUCCGCUCUCAUUGCCCCAUGUUCUCU__________ ....................((.(((((...((((.......))))........))))).))......................................................... ( -8.70 = -7.99 + -0.71)
| Location | 2,858,063 – 2,858,160 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 60.36 |
| Shannon entropy | 0.70643 |
| G+C content | 0.59096 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -12.84 |
| Energy contribution | -12.60 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.805831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
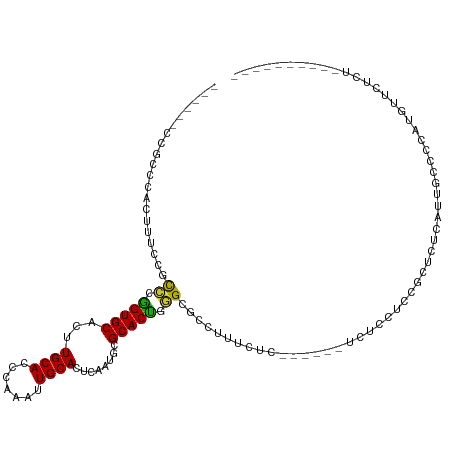
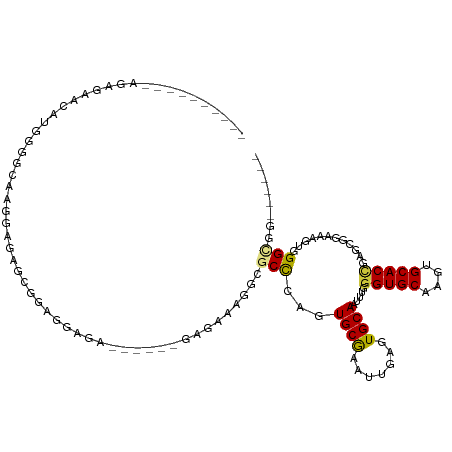
>dm3.chr2L 2858063 97 - 23011544 ----------AAAGAGCAUGGGGCGUGGAGAGCGGAGGAAA------GGGAAAGGCGCCCGGUGCAAAUUGAGUGCAAUUUGGGUGCAAGUGCACCGGGCGGAAAGUGGGCGG------ ----------.....((......(((.....))).......------........(((((((((((.......((((.......))))..)))))))))))........))..------ ( -31.30, z-score = -2.12, R) >droWil1.scaffold_180772 2961057 119 + 8906247 GUUUGGGGCCGGGGGAGGGCAGACAAUGAGAGCGAGAGAGACGCCAUGAGAAAAUGGCUUAGUGCGCGUUGGGUGCAAAUCAAGUGCAAAUGCACCUAAAGCACAGUGGGCUUAGGCUG (((((((.(((..(((..(((..(((((...(((..((....(((((......)))))))..))).)))))..)))...))..((((....))))........)..))).))))))).. ( -38.40, z-score = -1.15, R) >droPer1.super_10 34012 103 + 3432795 ----GAAGACUGAGAAAGAGAGGGAGUGACGGGAGGGGAGGCG---UUCGCUUGGCGCUGAGUGCGCGUUGAGUGCAAUUUAGGUGCAGCUGCACCGAGUCGAACGUAGG--------- ----....................................(((---((((((((...((((((((((.....)))).))))))((((....)))))))).)))))))...--------- ( -29.60, z-score = 0.24, R) >dp4.chr4_group4 2228882 103 + 6586962 ----GAAGACUGAGAAAGAGAGGGAGUGACGGGAGGGGAGGCG---UUCGCUUGGCGCUGAGUGCGCGUUGAGUGCAAUUUAGGUGCAGCUGCACCGAGUCGAACGUAGG--------- ----....................................(((---((((((((...((((((((((.....)))).))))))((((....)))))))).)))))))...--------- ( -29.60, z-score = 0.24, R) >droAna3.scaffold_12916 722891 96 - 16180835 -----------GAAAAGAGGGGGCAAAGGGGGGCAAAGACA-----CAAUUCGUUGGCCAAGUGCG-AUUGGGUGCAACAUGGGUGCACUUGCACUGGGCGUGUUGUGGGUGG------ -----------...........((........)).....((-----((((....((.(((.(((((-....((((((.......)))))))))))))).)).)))))).....------ ( -26.10, z-score = 0.01, R) >droSec1.super_5 1017125 97 - 5866729 ----------AGAGAGCAUGGGGCGAGGAGAGCGGAGGAGA------GAGAAAGGCGCCCGGUGCAAAUUGAGUGCAAUUUGGGUGCAAGUGCACCGUGCGGAAAGUGGGCGG------ ----------.....((((.(((((.......(........------).......))))).))))((((((....)))))).(((((....)))))((.((.....)).))..------ ( -30.74, z-score = -2.28, R) >droSim1.chr2L 2819058 97 - 22036055 ----------AGAGAGCAUGGGGCAAGGAGAGCGGAGGAGA------GAGAAAGGCGCCCGGUGCAAAUUGAGUGCAAUUUGGGUGCAAGUGCACCGGGCGGAAAGUGGGCGG------ ----------.....((.....((.......))........------........(((((((((((.......((((.......))))..)))))))))))........))..------ ( -30.40, z-score = -2.53, R) >consensus __________AGAGAACAUGGGGCAAGGAGAGCGGAGGAGA______GAGAAAGGCGCCCAGUGCGAAUUGAGUGCAAUUUGGGUGCAAGUGCACCGAGCGGAAAGUGGGCGG______ ........................................................(((...((((.......)))).....(((((....)))))............)))........ (-12.84 = -12.60 + -0.24)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:12:25 2011