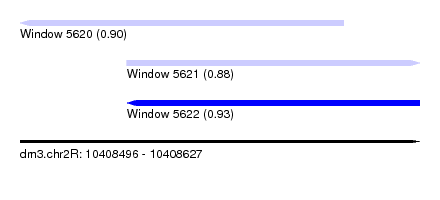
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,408,496 – 10,408,627 |
| Length | 131 |
| Max. P | 0.927735 |

| Location | 10,408,496 – 10,408,602 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 72.21 |
| Shannon entropy | 0.59629 |
| G+C content | 0.54775 |
| Mean single sequence MFE | -26.07 |
| Consensus MFE | -20.90 |
| Energy contribution | -21.00 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.13 |
| Mean z-score | -0.51 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.895668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10408496 106 - 21146708 UCUUCUGGCCGCACAUUUGCGGACGACGUGUGUCAACGCAUUGUGCGGUGUAAAUUGCUUGCUGCCAC--AUCUUCCCACACCCAAACCGAUUCCCGCAGCCCCGCAU-------- .......((((((((..((((...(((....)))..)))).))))))))......(((..(((((...--(((................)))....)))))...))).-------- ( -31.69, z-score = -1.39, R) >droAna3.scaffold_13266 10807678 115 - 19884421 UCUUCUGGCCGCACAUUUGCGGACGACGUGUGUCAACGCAUUGUGCGGUGUAAAUUGCUUGCUGCUAUCGACCUUAU-CCCCCUAUCCCCAUCCCCAUCUCCAUCCCCUGCCAUGC .....((((((((((..((((...(((....)))..)))).))))))..((..((.((.....)).))..)).....-...............................))))... ( -24.60, z-score = -0.59, R) >droEre2.scaffold_4845 7141927 92 + 22589142 UCUUCUGGCCGCACAUUUGCGGACGACGUGUGUCAACGCAUUGUGCGGUGUAAAUUGCUUGCUGCCAC--AUCUUGCCACACCCAUUCCCGCAU---------------------- .....((((.(((((..((((...(((....)))..)))).)))))(((((((.....)))).)))..--.....))))...............---------------------- ( -25.70, z-score = -0.13, R) >droYak2.chr2R 10335004 92 - 21139217 UCUUCUGGCCGCACAUUUGCGGACGACGUGUGUCAACGCAUUGUGCGAUGUAAAUUGCUUGCUGCCAC--AUCUUGCCACACCCAUCCUCGCAU---------------------- .....((((.(((((..((((...(((....)))..)))).)))))(((((.....((.....)).))--)))..))))...............---------------------- ( -25.40, z-score = -0.64, R) >droSec1.super_1 7919131 106 - 14215200 UCUUCUGGCCGCACAUUUGCGGACGACGUGUGUCAACGCAUUGUGCGGUGUAAAUUGCUUGCUGCCAU--AUCUUCCCACACCCAUGCCCAUUCCCGCAGCCCCGCAG-------- .......((((((((..((((...(((....)))..)))).))))))))......(((..(((((...--..........................)))))...))).-------- ( -30.65, z-score = -1.00, R) >droSim1.chr2R 9163089 106 - 19596830 UCUUCUGGCCGCACAUUUGCGGACGACGUGUGUCAACGCAUUGUGCGGUGUAAAUUGCUUGCUGCCAC--AUCUUCCCACACCCAUGCCCAUUCCCGCAGCCCCGCAG-------- .......((((((((..((((...(((....)))..)))).))))))))......(((..(((((...--..........................)))))...))).-------- ( -30.65, z-score = -0.85, R) >droWil1.scaffold_180701 968458 97 - 3904529 UCUUUUGGCCGCACAUUUGUGGACGACGUGUGUCAACGCAUUGUGCGGUGUAAAUUGCUUC-UACCCCACUACAAACGUCUCCGUCUCGUUCUCUUCU------------------ .......((((((((..((((...(((....)))..)))).))))))))............-............((((.........)))).......------------------ ( -21.30, z-score = -0.44, R) >droPer1.super_2 5236223 107 - 9036312 UCUUCUGGCCGCACAUUUGCGGACGACGUGUGUCAACGCAUUGUGCGGUGUAAAUUGCUUGCUUCUAUCAACCUUCCACCACCUAGCCCUCCCCUCCCCCACAUAGU--------- ......(((.(((((..((((...(((....)))..)))).)))))((((......(.(((.......))).)......))))..)))...................--------- ( -23.40, z-score = -0.77, R) >dp4.chr3 5036711 107 - 19779522 UCUUCUGGCCGCACAUUUGCGGACGACGUGUGUCAACGCAUUGUGCGGUGUAAAUUGCUUGCUUCUAUCAACCUUCCACCACCUAGCCCUCCCCUCCCCCACAUAGU--------- ......(((.(((((..((((...(((....)))..)))).)))))((((......(.(((.......))).)......))))..)))...................--------- ( -23.40, z-score = -0.77, R) >droVir3.scaffold_12875 10495288 98 + 20611582 UCCCGUAGCCGCACAUUUGCGGGCGACGUGUGUCAACGCAUUGUGCGGUGUAAAUUGCUUG-UCCUAGUCA--UUGCACCACCCAACUGCAAUGUACUCAG--------------- .......((((((((..((((...(((....)))..)))).))))))))............-..(((((((--(((((.........))))))).))).))--------------- ( -30.90, z-score = -1.07, R) >droMoj3.scaffold_6496 16908414 86 - 26866924 UCCCGUAGCCGCACAUUUGCGGACGACGUGUGUCAACGCAUUGUACGGUGUAAAUUGCUUG-UCCUACUCA--UUGCAACAGGC-CCCGC-------------------------- ..((((..(((((....)))))((((.((((....)))).))))))))........(((((-(..((....--.))..))))))-.....-------------------------- ( -22.80, z-score = 0.32, R) >droGri2.scaffold_15245 15048896 95 + 18325388 UCCCGUAGCCAUACAUUUGCGGGCGACGUGUGUCAACGCAUUGUGCGGUGUAAAUUGCUUG-CCCCACUCAACUUGCAACGACU-GCCACCACCGAC------------------- ..((((((........))))))(((..(((((....)))))..)))((((..(.(((.(((-(............))))))).)-..))))......------------------- ( -22.30, z-score = 1.17, R) >consensus UCUUCUGGCCGCACAUUUGCGGACGACGUGUGUCAACGCAUUGUGCGGUGUAAAUUGCUUGCUGCCAC__AUCUUCCAACACCCAUCCCCACUCCCCC__________________ .......((((((((..((((...(((....)))..)))).))))))))................................................................... (-20.90 = -21.00 + 0.10)


| Location | 10,408,531 – 10,408,627 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 74.63 |
| Shannon entropy | 0.49105 |
| G+C content | 0.52151 |
| Mean single sequence MFE | -25.05 |
| Consensus MFE | -18.38 |
| Energy contribution | -18.33 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.876276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10408531 96 + 21146708 -----GAU--GUGGCAGCAAGCAAUUUACACCGCACAAUGCGUUGACACACGUCGUCCGCAAAUGUGCGGCCAGAAGACAAUAGAGAAAUAGAGGCAGCCAGG----------- -----...--.((((.((...(.((((...(((((((.((((.((((....))))..))))..)))))))........(......))))).)..)).))))..----------- ( -26.80, z-score = -1.41, R) >droAna3.scaffold_13266 10807715 98 + 19884421 GAUAAGGUCGAUAGCAGCAAGCAAUUUACACCGCACAAUGCGUUGACACACGUCGUCCGCAAAUGUGCGGCCAGAAGACAAUAGAGAAAUAGGG--AGCC-------------- ......(((....((.....))........(((((((.((((.((((....))))..))))..)))))))......)))...............--....-------------- ( -24.60, z-score = -1.38, R) >droEre2.scaffold_4845 7141948 94 - 22589142 -----GAU--GUGGCAGCAAGCAAUUUACACCGCACAAUGCGUUGACACACGUCGUCCGCAAAUGUGCGGCCAGAAGACAAUAGAGAAAUAGGG--AGCCCGG----------- -----..(--.((((.(....).........((((((.((((.((((....))))..))))..)))))))))).)................((.--...))..----------- ( -25.30, z-score = -0.62, R) >droYak2.chr2R 10335025 92 + 21139217 -----GAU--GUGGCAGCAAGCAAUUUACAUCGCACAAUGCGUUGACACACGUCGUCCGCAAAUGUGCGGCCAGAAGACAAUAGAGAAAUAGGG--AGCCU------------- -----(((--(((((.....))....))))))(((((.((((.((((....))))..))))..)))))((((.....................)--.))).------------- ( -22.70, z-score = -0.10, R) >droSec1.super_1 7919166 96 + 14215200 -----GAU--AUGGCAGCAAGCAAUUUACACCGCACAAUGCGUUGACACACGUCGUCCGCAAAUGUGCGGCCAGAAGACAAUAGAGAAAUAGAGGCAGCCAGG----------- -----...--.((((.((...(.((((...(((((((.((((.((((....))))..))))..)))))))........(......))))).)..)).))))..----------- ( -26.90, z-score = -2.09, R) >droSim1.chr2R 9163124 96 + 19596830 -----GAU--GUGGCAGCAAGCAAUUUACACCGCACAAUGCGUUGACACACGUCGUCCGCAAAUGUGCGGCCAGAAGACAAUAGAGAAAUAGAGGCAGCCAGG----------- -----...--.((((.((...(.((((...(((((((.((((.((((....))))..))))..)))))))........(......))))).)..)).))))..----------- ( -26.80, z-score = -1.41, R) >droWil1.scaffold_180701 968490 85 + 3904529 ------------GGUAG-AAGCAAUUUACACCGCACAAUGCGUUGACACACGUCGUCCACAAAUGUGCGGCCAAAAGACAAUGGCGUGCCUCAACCAG---------------- ------------(((.(-(.(((........((((((.(..((.(((.......))).)).).))))))((((........)))).))).)).)))..---------------- ( -26.00, z-score = -2.01, R) >droPer1.super_2 5236257 109 + 9036312 -----GGUUGAUAGAAGCAAGCAAUUUACACCGCACAAUGCGUUGACACACGUCGUCCGCAAAUGUGCGGCCAGAAGACAAUAGAGCCAGGCAGCCAGACACCCACCCAGAGAG -----(((((......((((....))....(((((((.((((.((((....))))..))))..)))))))...............))....))))).................. ( -26.10, z-score = -1.46, R) >dp4.chr3 5036745 109 + 19779522 -----GGUUGAUAGAAGCAAGCAAUUUACACCGCACAAUGCGUUGACACACGUCGUCCGCAAAUGUGCGGCCAGAAGACAAUAGAGCCAGGCAGCCAGACACCCACCCAGAGAG -----(((((......((((....))....(((((((.((((.((((....))))..))))..)))))))...............))....))))).................. ( -26.10, z-score = -1.46, R) >droVir3.scaffold_12875 10495316 87 - 20611582 --------UGACUAGGACAAGCAAUUUACACCGCACAAUGCGUUGACACACGUCGCCCGCAAAUGUGCGGCUACGGGA-AGCC-C-AAAGACAAUGGG---------------- --------...(((................(((((((.((((.((((....))))..))))..)))))))....(((.-..))-)-........))).---------------- ( -23.10, z-score = -0.49, R) >droMoj3.scaffold_6496 16908430 89 + 26866924 --------UGAGUAGGACAAGCAAUUUACACCGUACAAUGCGUUGACACACGUCGUCCGCAAAUGUGCGGCUACGGGA-AGAGACCAAAGACAAUGGU---------------- --------...((((...((....))....(((((((.((((.((((....))))..))))..)))))))))))....-....((((.......))))---------------- ( -21.90, z-score = -0.30, R) >droGri2.scaffold_15245 15048919 89 - 18325388 ------GUUGAGUGGGGCAAGCAAUUUACACCGCACAAUGCGUUGACACACGUCGCCCGCAAAUGUAUGGCUACGGGA-AGCC-CGAAAGACAAUGG----------------- ------(((...(.((((..........((.(((.....))).)).....(((.(((.((....))..))).)))...-.)))-).)..))).....----------------- ( -24.30, z-score = 0.21, R) >consensus _____GAU__AUGGCAGCAAGCAAUUUACACCGCACAAUGCGUUGACACACGUCGUCCGCAAAUGUGCGGCCAGAAGACAAUAGAGAAAUACAG_CAGCC______________ ..............................(((((((.((((.((((....))))..))))..)))))))............................................ (-18.38 = -18.33 + -0.05)

| Location | 10,408,531 – 10,408,627 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 74.63 |
| Shannon entropy | 0.49105 |
| G+C content | 0.52151 |
| Mean single sequence MFE | -28.33 |
| Consensus MFE | -20.90 |
| Energy contribution | -21.00 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.927735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10408531 96 - 21146708 -----------CCUGGCUGCCUCUAUUUCUCUAUUGUCUUCUGGCCGCACAUUUGCGGACGACGUGUGUCAACGCAUUGUGCGGUGUAAAUUGCUUGCUGCCAC--AUC----- -----------..((((.((.......................((((((((..((((...(((....)))..)))).))))))))((.....))..)).)))).--...----- ( -29.40, z-score = -1.87, R) >droAna3.scaffold_13266 10807715 98 - 19884421 --------------GGCU--CCCUAUUUCUCUAUUGUCUUCUGGCCGCACAUUUGCGGACGACGUGUGUCAACGCAUUGUGCGGUGUAAAUUGCUUGCUGCUAUCGACCUUAUC --------------....--...............(((.....((((((((..((((...(((....)))..)))).)))))))).......((.....))....)))...... ( -26.00, z-score = -1.25, R) >droEre2.scaffold_4845 7141948 94 + 22589142 -----------CCGGGCU--CCCUAUUUCUCUAUUGUCUUCUGGCCGCACAUUUGCGGACGACGUGUGUCAACGCAUUGUGCGGUGUAAAUUGCUUGCUGCCAC--AUC----- -----------...(((.--.......................((((((((..((((...(((....)))..)))).))))))))((((.....)))).)))..--...----- ( -28.20, z-score = -1.22, R) >droYak2.chr2R 10335025 92 - 21139217 -------------AGGCU--CCCUAUUUCUCUAUUGUCUUCUGGCCGCACAUUUGCGGACGACGUGUGUCAACGCAUUGUGCGAUGUAAAUUGCUUGCUGCCAC--AUC----- -------------.(((.--...............(((....)))((((((..((((...(((....)))..)))).))))))..((((.....)))).)))..--...----- ( -24.20, z-score = -0.72, R) >droSec1.super_1 7919166 96 - 14215200 -----------CCUGGCUGCCUCUAUUUCUCUAUUGUCUUCUGGCCGCACAUUUGCGGACGACGUGUGUCAACGCAUUGUGCGGUGUAAAUUGCUUGCUGCCAU--AUC----- -----------..((((.((.......................((((((((..((((...(((....)))..)))).))))))))((.....))..)).)))).--...----- ( -29.50, z-score = -2.14, R) >droSim1.chr2R 9163124 96 - 19596830 -----------CCUGGCUGCCUCUAUUUCUCUAUUGUCUUCUGGCCGCACAUUUGCGGACGACGUGUGUCAACGCAUUGUGCGGUGUAAAUUGCUUGCUGCCAC--AUC----- -----------..((((.((.......................((((((((..((((...(((....)))..)))).))))))))((.....))..)).)))).--...----- ( -29.40, z-score = -1.87, R) >droWil1.scaffold_180701 968490 85 - 3904529 ----------------CUGGUUGAGGCACGCCAUUGUCUUUUGGCCGCACAUUUGUGGACGACGUGUGUCAACGCAUUGUGCGGUGUAAAUUGCUU-CUACC------------ ----------------..(((.((((((.((((........))))((((((..((((...(((....)))..)))).))))))........)))))-).)))------------ ( -32.00, z-score = -2.40, R) >droPer1.super_2 5236257 109 - 9036312 CUCUCUGGGUGGGUGUCUGGCUGCCUGGCUCUAUUGUCUUCUGGCCGCACAUUUGCGGACGACGUGUGUCAACGCAUUGUGCGGUGUAAAUUGCUUGCUUCUAUCAACC----- .......((((((.((..(((.....(((......))).....((((((((..((((...(((....)))..)))).)))))))).......))).)).))))))....----- ( -34.40, z-score = -1.10, R) >dp4.chr3 5036745 109 - 19779522 CUCUCUGGGUGGGUGUCUGGCUGCCUGGCUCUAUUGUCUUCUGGCCGCACAUUUGCGGACGACGUGUGUCAACGCAUUGUGCGGUGUAAAUUGCUUGCUUCUAUCAACC----- .......((((((.((..(((.....(((......))).....((((((((..((((...(((....)))..)))).)))))))).......))).)).))))))....----- ( -34.40, z-score = -1.10, R) >droVir3.scaffold_12875 10495316 87 + 20611582 ----------------CCCAUUGUCUUU-G-GGCU-UCCCGUAGCCGCACAUUUGCGGGCGACGUGUGUCAACGCAUUGUGCGGUGUAAAUUGCUUGUCCUAGUCA-------- ----------------.....((.((..-(-(((.-.......((((((((..((((...(((....)))..)))).))))))))...........)))).)).))-------- ( -28.41, z-score = -1.04, R) >droMoj3.scaffold_6496 16908430 89 - 26866924 ----------------ACCAUUGUCUUUGGUCUCU-UCCCGUAGCCGCACAUUUGCGGACGACGUGUGUCAACGCAUUGUACGGUGUAAAUUGCUUGUCCUACUCA-------- ----------------((((.......))))....-..((((..(((((....)))))((((.((((....)))).))))))))......................-------- ( -20.40, z-score = 0.12, R) >droGri2.scaffold_15245 15048919 89 + 18325388 -----------------CCAUUGUCUUUCG-GGCU-UCCCGUAGCCAUACAUUUGCGGGCGACGUGUGUCAACGCAUUGUGCGGUGUAAAUUGCUUGCCCCACUCAAC------ -----------------............(-(((.-..((((((........))))))((((.((((....)))).))))(((((....)))))..))))........------ ( -23.60, z-score = 0.36, R) >consensus ______________GGCUG_CUCUAUUUCUCUAUUGUCUUCUGGCCGCACAUUUGCGGACGACGUGUGUCAACGCAUUGUGCGGUGUAAAUUGCUUGCUGCCAU__AUC_____ ...........................................((((((((..((((...(((....)))..)))).))))))))............................. (-20.90 = -21.00 + 0.10)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:22:47 2011