| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,399,158 – 10,399,248 |
| Length | 90 |
| Max. P | 0.993974 |

| Location | 10,399,158 – 10,399,248 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 79.70 |
| Shannon entropy | 0.41016 |
| G+C content | 0.32322 |
| Mean single sequence MFE | -20.67 |
| Consensus MFE | -15.71 |
| Energy contribution | -15.72 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.990704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
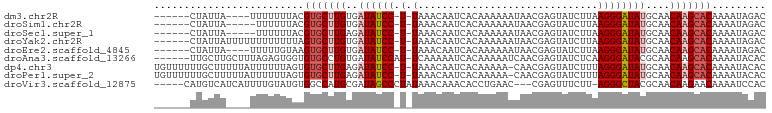
>dm3.chr2R 10399158 90 + 21146708 GUCUAUUUUGUGCUUGUUGCAUAUCCCUUAAGAUACUCGUUAUUUUUUGUGAUUGUUUA-A-GGAUAUCACAAGCACGUAAAAAAA----UAAUAG------ .........((((((((.(.((((((.(((((......(((((.....)))))..))))-)-))))))))))))))).........----......------ ( -20.50, z-score = -3.00, R) >droSim1.chr2R 9153626 89 + 19596830 GUCUAUUUUGUGCUUGUUGCAUAUCCCUUAAGAUACUCGUUAUUUUUUGUGAUUGUUUA-A-GGAUAUCACAAGCACGUAAAAAA-----UAAUAG------ .........((((((((.(.((((((.(((((......(((((.....)))))..))))-)-)))))))))))))))........-----......------ ( -20.50, z-score = -3.05, R) >droSec1.super_1 7909701 89 + 14215200 GUCUAUUUUGUGCUUGUUGCAUAUCCCUUAAGAUACUCGUUAUUUUUUGUGAUUGUUUA-A-GGAUAUCUCAAGCACGUAAAAAA-----UAAUAG------ .........(((((((..(.((((((.(((((......(((((.....)))))..))))-)-))))))).)))))))........-----......------ ( -18.80, z-score = -2.68, R) >droYak2.chr2R 10324805 94 + 21139217 GUCUAUUUUGUGCUUGUUGCAUAUCCCUUAAGAUACUCGUUAUUUUUUGUGAUUGUUUA-A-GGAUAUCACAAGCACUAAAAAAAAAAAAUAAUAG------ .....((((((((((((.(.((((((.(((((......(((((.....)))))..))))-)-))))))))))))))).))))..............------ ( -20.40, z-score = -3.19, R) >droEre2.scaffold_4845 7132569 90 - 22589142 GUCUAUUUUGUGCUUGUUGCAUAUCCCUUAAGAUACUCGUUAUUUUUUGUGAUUGUUUA-A-GGAUAUCACAAGCACUUACAAAAA----UAAUAG------ .........((((((((.(.((((((.(((((......(((((.....)))))..))))-)-))))))))))))))).........----......------ ( -20.00, z-score = -2.87, R) >droAna3.scaffold_13266 14723843 95 + 19884421 GUGUAUUUUGUGCUUGUUGCGUAUCCCUUGAGAUACUCGUUGAUUUUUGUGAUUUUUGA-AUGGAUAUCACAGGCACACCACUCUAAAGCAAGCAA------ ..........(((((((((.(((((......))))).).......((((((((.(((..-..))).)))))))).............)))))))).------ ( -21.00, z-score = -0.57, R) >dp4.chr3 5027182 99 + 19779522 GUGUAUUUUGUGCUUGUUGCAUAUCCCUAAAGAUACUCGUUG-UUUUUGUGAUUGUUUA-A-GGAUAUCUCAAGCACACUAAAAAAUAAAAAGCAAAAAACA ((.(((((((((((((..(.((((((.((((.(....((...-....))....).))))-.-))))))).)))))))).....)))))....))........ ( -19.80, z-score = -1.70, R) >droPer1.super_2 5226555 99 + 9036312 GUGUAUUUUGUGCUUGUUGCAUAUCCCUAAAGAUACUCGUUG-UUUUUGUGAUUGUUUA-A-GGAUAUCUCAAGCACACUAAAAAAUAAAAAGCAAAAAACA ((.(((((((((((((..(.((((((.((((.(....((...-....))....).))))-.-))))))).)))))))).....)))))....))........ ( -19.80, z-score = -1.70, R) >droVir3.scaffold_12875 13778024 93 - 20611582 GUGGAUUUUGUUCUUGUUGCGUAGCCCU-AAGAAACUCG---GUUCAGGUGUUUGUUUAUAGGGCUAUCGCAUGGCCACAUACAAAAUGAUGACAUG----- ....(((((((...((((((((((((((-((.((((((.---.....)).)))).)...)))))))).)))).....))).))))))).........----- ( -25.20, z-score = -1.17, R) >consensus GUCUAUUUUGUGCUUGUUGCAUAUCCCUUAAGAUACUCGUUAUUUUUUGUGAUUGUUUA_A_GGAUAUCACAAGCACACAAAAAAA____UAAUAG______ .........(((((((....((((((....(((((.(((..........))).)))))....))))))..)))))))......................... (-15.71 = -15.72 + 0.02)


| Location | 10,399,158 – 10,399,248 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 79.70 |
| Shannon entropy | 0.41016 |
| G+C content | 0.32322 |
| Mean single sequence MFE | -19.81 |
| Consensus MFE | -12.16 |
| Energy contribution | -12.74 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.993974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
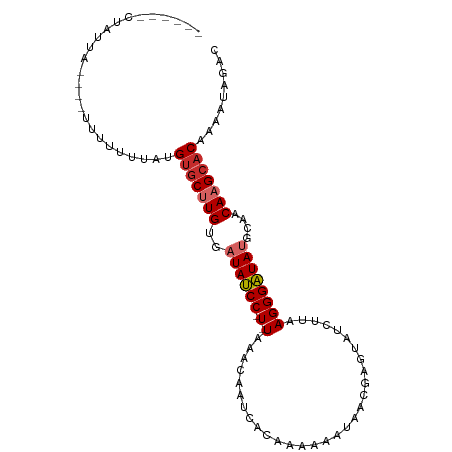
>dm3.chr2R 10399158 90 - 21146708 ------CUAUUA----UUUUUUUACGUGCUUGUGAUAUCC-U-UAAACAAUCACAAAAAAUAACGAGUAUCUUAAGGGAUAUGCAACAAGCACAAAAUAGAC ------......----.........((((((((.((((((-(-(......((............))........))))))))...))))))))......... ( -18.94, z-score = -3.38, R) >droSim1.chr2R 9153626 89 - 19596830 ------CUAUUA-----UUUUUUACGUGCUUGUGAUAUCC-U-UAAACAAUCACAAAAAAUAACGAGUAUCUUAAGGGAUAUGCAACAAGCACAAAAUAGAC ------...(((-----((((....((((((((.((((((-(-(......((............))........))))))))...))))))))))))))).. ( -19.14, z-score = -3.58, R) >droSec1.super_1 7909701 89 - 14215200 ------CUAUUA-----UUUUUUACGUGCUUGAGAUAUCC-U-UAAACAAUCACAAAAAAUAACGAGUAUCUUAAGGGAUAUGCAACAAGCACAAAAUAGAC ------...(((-----((((....(((((((..((((((-(-(......((............))........))))))))....)))))))))))))).. ( -16.24, z-score = -2.79, R) >droYak2.chr2R 10324805 94 - 21139217 ------CUAUUAUUUUUUUUUUUUAGUGCUUGUGAUAUCC-U-UAAACAAUCACAAAAAAUAACGAGUAUCUUAAGGGAUAUGCAACAAGCACAAAAUAGAC ------...................((((((((.((((((-(-(......((............))........))))))))...))))))))......... ( -18.84, z-score = -2.91, R) >droEre2.scaffold_4845 7132569 90 + 22589142 ------CUAUUA----UUUUUGUAAGUGCUUGUGAUAUCC-U-UAAACAAUCACAAAAAAUAACGAGUAUCUUAAGGGAUAUGCAACAAGCACAAAAUAGAC ------......----.........((((((((.((((((-(-(......((............))........))))))))...))))))))......... ( -18.84, z-score = -2.85, R) >droAna3.scaffold_13266 14723843 95 - 19884421 ------UUGCUUGCUUUAGAGUGGUGUGCCUGUGAUAUCCAU-UCAAAAAUCACAAAAAUCAACGAGUAUCUCAAGGGAUACGCAACAAGCACAAAAUACAC ------.((((((.....(((((((((.......))).))))-))...................(.((((((....)))))).)..)))))).......... ( -21.70, z-score = -1.89, R) >dp4.chr3 5027182 99 - 19779522 UGUUUUUUGCUUUUUAUUUUUUAGUGUGCUUGAGAUAUCC-U-UAAACAAUCACAAAAA-CAACGAGUAUCUUUAGGGAUAUGCAACAAGCACAAAAUACAC ........................((((((((..((((((-(-((((...((.......-....)).....)))))))))))....))))))))........ ( -20.40, z-score = -2.28, R) >droPer1.super_2 5226555 99 - 9036312 UGUUUUUUGCUUUUUAUUUUUUAGUGUGCUUGAGAUAUCC-U-UAAACAAUCACAAAAA-CAACGAGUAUCUUUAGGGAUAUGCAACAAGCACAAAAUACAC ........................((((((((..((((((-(-((((...((.......-....)).....)))))))))))....))))))))........ ( -20.40, z-score = -2.28, R) >droVir3.scaffold_12875 13778024 93 + 20611582 -----CAUGUCAUCAUUUUGUAUGUGGCCAUGCGAUAGCCCUAUAAACAAACACCUGAAC---CGAGUUUCUU-AGGGCUACGCAACAAGAACAAAAUCCAC -----.........(((((((...((....((((.((((((((.((((............---...))))..)-))))))))))).))...))))))).... ( -23.76, z-score = -2.81, R) >consensus ______CUAUUA____UUUUUUUAUGUGCUUGUGAUAUCC_U_UAAACAAUCACAAAAAAUAACGAGUAUCUUAAGGGAUAUGCAACAAGCACAAAAUAGAC .........................(((((((..((((((..........((............))..........))))))....)))))))......... (-12.16 = -12.74 + 0.58)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:22:44 2011