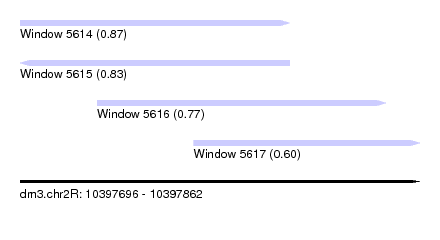
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,397,696 – 10,397,862 |
| Length | 166 |
| Max. P | 0.866732 |

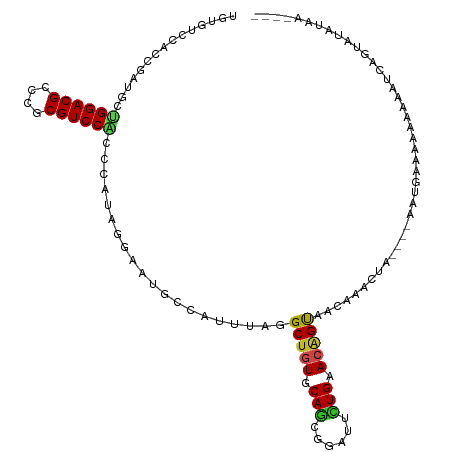
| Location | 10,397,696 – 10,397,808 |
|---|---|
| Length | 112 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 72.99 |
| Shannon entropy | 0.58193 |
| G+C content | 0.49478 |
| Mean single sequence MFE | -33.66 |
| Consensus MFE | -18.69 |
| Energy contribution | -18.71 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.866732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10397696 112 + 21146708 UUAGUUGCAUACUAAUAUUUGUGCCAUU----UAGUUCGUUACUGUUCAGAAUCCGCUGCACAGCAUAAACGGCAUUCCUAUGGGUGGACGCGGGCGUCCAGCAUCGGUGGACACA (((((.....)))))....(((((((((----..((.((((.((((.(((......))).))))....)))))).........(.((((((....)))))).)...))))).)))) ( -31.00, z-score = -0.20, R) >droSim1.chr2R 9152169 108 + 19596830 ----UUACAUGCUAAUAUUUGUAUCAUU----UAGUUUGUUACUGUUCAGAAUCCGCUGCACAGCAUAAACGGCAUUCCUAUGGGUGGACGCGGGCGUCCAGCAUCGGUGGACACA ----....(((((..((..((...))..----))((((((..((((.(((......))).)))).)))))))))))(((....(.((((((....)))))).)......))).... ( -27.00, z-score = 0.15, R) >droSec1.super_1 7908219 108 + 14215200 ----UUACAUACUAAUAUUUGUGCCAUU----UAGUUUGUUACUGUUCAGAAUCCUCUGCACAGCAUAAACGGCAUUCCUAGGUGUGGACGGGGGCGUCCAGCAUCGGUGGACACA ----...............(((((((((----..((((((..((((.((((....)))).)))).))))))..........((((((((((....)))))).))))))))).)))) ( -33.60, z-score = -1.90, R) >droYak2.chr2R 10323345 108 + 21139217 ----UUGUAUACUCAUUUUUGUGCCAUU----UAGUUCGUUACUGUUCAAAAUCCGCUGCACAGCAUAAACGGCAUUCCUAUUGGUGGACGCGGGCGUCCAGCAUCGGUGGACACA ----.(((.((((.......(((((...----((((.....))))..........(((....)))......)))))......((.((((((....)))))).))..)))).))).. ( -27.30, z-score = -0.03, R) >droEre2.scaffold_4845 7131114 108 - 22589142 ----UUACAUACUCAUUUUUGUGUCAUU----UAGUUCGUUACUGUUCAAAAUCCGCUGCACAGCAUAAACGGCAUUCCUAUUGGUGGACGCGGGCGUCCAGCAUCGGUGGACACA ----...............((((((...----..((.((((.((((.((........)).))))....))))))...((...((.((((((....)))))).))..))..)))))) ( -27.70, z-score = -0.89, R) >droAna3.scaffold_13266 14722554 112 + 19884421 ----UGAAGCCUACAACCUCUUUGCCUUGCAGCAGUUCGUCACUGUUCAGAAUCCACUGCACAGCCUCAACGGCAUAUCAAUGAGUGGACGCGGGCGUCCAGCGUCGGUGGAUACA ----......................(((.((((((.....))))))))).((((((((.((.(((.....)))...........((((((....))))))..))))))))))... ( -33.10, z-score = -0.00, R) >dp4.chr3 5025680 109 + 19779522 ----GCAUCUAAUGAUU-AUUGUGCAUUGG--CAGUUUGUCACCGUUCAGAAUCCGCUGCACAGCCUGAAUGGCCUGCCAAUGGGCGGACGCGGGCGUCCCGCAUCGGUGGAUAAU ----..(((((.((((.-.((((.((((((--(((.......((((((((.....(((....))))))))))).))))))))).))))..(((((...))))))))).)))))... ( -42.80, z-score = -1.59, R) >droPer1.super_2 5225048 109 + 9036312 ----AUAUCUAAUGAUU-AUUGUGCAUUGG--CAGUUUGUCACCGUUCAGAAUCCGCUGCACAGCCUGAAUGGCCUGCCAAUGGGCGGACGCGGGCGUCCCGCAUCGGUGGAUAAU ----.((((((.((((.-.((((.((((((--(((.......((((((((.....(((....))))))))))).))))))))).))))..(((((...))))))))).)))))).. ( -44.20, z-score = -2.34, R) >droWil1.scaffold_180701 950001 110 + 3904529 ----GAAUUUAAUGAUUUGAAAUUUGUUUU--UAGUUUGUCACUGUGCAAAAUCCAUUGCAUAGCCUUAAUGGUCUACCAAUGGGCGGACGCGGGCGUCCAGCUUCGGUGGAUAAU ----........((((((((((.....)))--)))...))))((((((((......))))))))........(((((((...(((((((((....))))).)))).)))))))... ( -32.30, z-score = -1.82, R) >droVir3.scaffold_12875 13776037 101 - 20611582 -------CUAAUUGAAUUGCUUUACA--------GUUUGUCGCCGUGCAGAAUCCGCUGCACAGCCUCAAUGGCAUAAGCAUGGGCGGACGCGGGCGUCCGGCUUCUGUAGACAAU -------........((((.((((((--------(...((((((((((((......)))))).(((.....))).........)))(((((....))))))))..))))))))))) ( -42.10, z-score = -3.22, R) >droMoj3.scaffold_6496 13396173 104 + 26866924 -------UCAAAUUAAUUCCUUUUCAUC-----AGUUUGUUGCCGUUCAGAAUUCGCUGCACAGCCUUAAUGGCAUUAGCUUGGGCGGACGCGGGCGUCCGGCUUCAGUGGACAAC -------.....................-----.(((..(((((((.(((......))).)).((((....(((....))).))))(((((....))))))))...))..)))... ( -29.60, z-score = -0.64, R) >droGri2.scaffold_15112 3468731 107 - 5172618 ---------UGAUUAAUCCCUUUUUACUGGCAUAGUUUGUCACUGUGCAGAAUCCGUUGCAUGGCUUGAAUGGCAUUACGAUGGGCGGACGCGGGCGUCCGGCUUCUGUGGACAAC ---------.......(((..........(((((((.....)))))))(((((((((((....(((.....)))....)))))))((((((....))))))..))))..))).... ( -33.20, z-score = -0.61, R) >consensus ____UUACAUAAUGAUUUUUUUUUCAUU____UAGUUUGUCACUGUUCAGAAUCCGCUGCACAGCCUAAACGGCAUUCCUAUGGGCGGACGCGGGCGUCCAGCAUCGGUGGACAAA ....................................................(((((((....((.......)).........(.((((((....)))))).)..))))))).... (-18.69 = -18.71 + 0.02)

| Location | 10,397,696 – 10,397,808 |
|---|---|
| Length | 112 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 72.99 |
| Shannon entropy | 0.58193 |
| G+C content | 0.49478 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -15.72 |
| Energy contribution | -15.34 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.833470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10397696 112 - 21146708 UGUGUCCACCGAUGCUGGACGCCCGCGUCCACCCAUAGGAAUGCCGUUUAUGCUGUGCAGCGGAUUCUGAACAGUAACGAACUA----AAUGGCACAAAUAUUAGUAUGCAACUAA (((((.(.((.(((.((((((....))))))..))).))..(((((((((((((((.(((......))).))))))......))----))))))).........).)))))..... ( -32.80, z-score = -1.57, R) >droSim1.chr2R 9152169 108 - 19596830 UGUGUCCACCGAUGCUGGACGCCCGCGUCCACCCAUAGGAAUGCCGUUUAUGCUGUGCAGCGGAUUCUGAACAGUAACAAACUA----AAUGAUACAAAUAUUAGCAUGUAA---- ((((((..((.(((.((((((....))))))..))).))......((((.((((((.(((......))).))))))..))))..----...))))))...............---- ( -28.40, z-score = -1.18, R) >droSec1.super_1 7908219 108 - 14215200 UGUGUCCACCGAUGCUGGACGCCCCCGUCCACACCUAGGAAUGCCGUUUAUGCUGUGCAGAGGAUUCUGAACAGUAACAAACUA----AAUGGCACAAAUAUUAGUAUGUAA---- .(((((((.......))))))).....(((.......))).(((((((((((((((.(((......))).))))))......))----))))))).................---- ( -30.10, z-score = -1.80, R) >droYak2.chr2R 10323345 108 - 21139217 UGUGUCCACCGAUGCUGGACGCCCGCGUCCACCAAUAGGAAUGCCGUUUAUGCUGUGCAGCGGAUUUUGAACAGUAACGAACUA----AAUGGCACAAAAAUGAGUAUACAA---- (((((.(.((..((.((((((....)))))).))...))..(((((((((((((((.(((......))).))))))......))----))))))).........).))))).---- ( -32.30, z-score = -1.95, R) >droEre2.scaffold_4845 7131114 108 + 22589142 UGUGUCCACCGAUGCUGGACGCCCGCGUCCACCAAUAGGAAUGCCGUUUAUGCUGUGCAGCGGAUUUUGAACAGUAACGAACUA----AAUGACACAAAAAUGAGUAUGUAA---- ((((((..((..((.((((((....)))))).))...))......((((.((((((.(((......))).))))))..))))..----...))))))...............---- ( -29.20, z-score = -1.47, R) >droAna3.scaffold_13266 14722554 112 - 19884421 UGUAUCCACCGACGCUGGACGCCCGCGUCCACUCAUUGAUAUGCCGUUGAGGCUGUGCAGUGGAUUCUGAACAGUGACGAACUGCUGCAAGGCAAAGAGGUUGUAGGCUUCA---- .....((..((((..((((((....))))))(((.(((....(((.....)))..(((((..(.(((...........))))..)))))...))).)))))))..)).....---- ( -34.90, z-score = 0.22, R) >dp4.chr3 5025680 109 - 19779522 AUUAUCCACCGAUGCGGGACGCCCGCGUCCGCCCAUUGGCAGGCCAUUCAGGCUGUGCAGCGGAUUCUGAACGGUGACAAACUG--CCAAUGCACAAU-AAUCAUUAGAUGC---- ..((((....((((((((...)))))))).(..((((((((((((.(((((.(((.....)))...))))).)))......)))--))))))..)...-........)))).---- ( -38.00, z-score = -1.42, R) >droPer1.super_2 5225048 109 - 9036312 AUUAUCCACCGAUGCGGGACGCCCGCGUCCGCCCAUUGGCAGGCCAUUCAGGCUGUGCAGCGGAUUCUGAACGGUGACAAACUG--CCAAUGCACAAU-AAUCAUUAGAUAU---- ..((((....((((((((...)))))))).(..((((((((((((.(((((.(((.....)))...))))).)))......)))--))))))..)...-........)))).---- ( -38.40, z-score = -1.92, R) >droWil1.scaffold_180701 950001 110 - 3904529 AUUAUCCACCGAAGCUGGACGCCCGCGUCCGCCCAUUGGUAGACCAUUAAGGCUAUGCAAUGGAUUUUGCACAGUGACAAACUA--AAAACAAAUUUCAAAUCAUUAAAUUC---- ..........(((((.(((((....)))))..((((((((((.((.....)))))).)))))).....))..(((.....))).--.........)))..............---- ( -24.70, z-score = -1.59, R) >droVir3.scaffold_12875 13776037 101 + 20611582 AUUGUCUACAGAAGCCGGACGCCCGCGUCCGCCCAUGCUUAUGCCAUUGAGGCUGUGCAGCGGAUUCUGCACGGCGACAAAC--------UGUAAAGCAAUUCAAUUAG------- (((((.(((((((((((((((....)))))).....))))......(((..(((((((((......)))))))))..))).)--------))))..)))))........------- ( -36.00, z-score = -2.35, R) >droMoj3.scaffold_6496 13396173 104 - 26866924 GUUGUCCACUGAAGCCGGACGCCCGCGUCCGCCCAAGCUAAUGCCAUUAAGGCUGUGCAGCGAAUUCUGAACGGCAACAAACU-----GAUGAAAAGGAAUUAAUUUGA------- (((((.(((((.(((((((((....)))))......((....))......))))...))).......)).)))))..((((.(-----(((........)))).)))).------- ( -26.21, z-score = -0.27, R) >droGri2.scaffold_15112 3468731 107 + 5172618 GUUGUCCACAGAAGCCGGACGCCCGCGUCCGCCCAUCGUAAUGCCAUUCAAGCCAUGCAACGGAUUCUGCACAGUGACAAACUAUGCCAGUAAAAAGGGAUUAAUCA--------- ...((((..((((.(((((((....))))).......(((..((.......))..)))...)).))))(((.(((.....))).)))..........))))......--------- ( -24.60, z-score = 0.10, R) >consensus UGUGUCCACCGAUGCUGGACGCCCGCGUCCACCCAUAGGAAUGCCAUUUAGGCUGUGCAGCGGAUUCUGAACAGUAACAAACUA____AAUGAAAAAAAAAUCAGUAUAUAA____ ...............((((((....))))))....................(((((.(((......))).)))))......................................... (-15.72 = -15.34 + -0.38)
| Location | 10,397,728 – 10,397,848 |
|---|---|
| Length | 120 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.62 |
| Shannon entropy | 0.55740 |
| G+C content | 0.55867 |
| Mean single sequence MFE | -41.31 |
| Consensus MFE | -23.06 |
| Energy contribution | -22.38 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.766148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
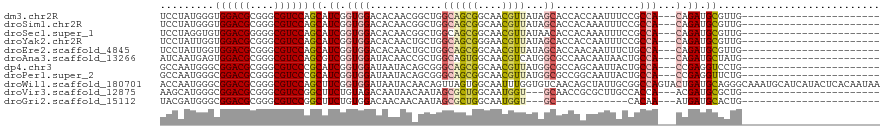
>dm3.chr2R 10397728 120 + 21146708 UCGUUACUGUUCAGAAUCCGCUGCACAGCAUAAACGGCAUUCCUAUGGGUGGACGCGGGCGUCCAGCAUCGGUGGACACAACGGCUGGCAGCGGCAACGUUAUAGCACCACCAAUUUCCG .((((.((((.(((......))).))))....))))((...((....))((((((....))))))))...(((((.(.....)((((..((((....)))).)))).)))))........ ( -39.10, z-score = -0.79, R) >droSim1.chr2R 9152197 120 + 19596830 UUGUUACUGUUCAGAAUCCGCUGCACAGCAUAAACGGCAUUCCUAUGGGUGGACGCGGGCGUCCAGCAUCGGUGGACACAACGGCUGGCAGCGGCAACGUUAUAGCACCACAAAUUUCCG ((((..((((.......(((((((.((((.......((...((....))((((((....))))))))..((.((....)).))))))))))))).......))))....))))....... ( -38.24, z-score = -0.69, R) >droSec1.super_1 7908247 120 + 14215200 UUGUUACUGUUCAGAAUCCUCUGCACAGCAUAAACGGCAUUCCUAGGUGUGGACGGGGGCGUCCAGCAUCGGUGGACACAACGGCUGGCAGCGGCAACGUUAUAACACCACAAAUUUCCG ......((((.((((....)))).))))......(((........(((((.((((..(.((((((((....((....))....)))))..))).)..))))...)))))........))) ( -38.59, z-score = -1.42, R) >droYak2.chr2R 10323373 120 + 21139217 UCGUUACUGUUCAAAAUCCGCUGCACAGCAUAAACGGCAUUCCUAUUGGUGGACGCGGGCGUCCAGCAUCGGUGGACACAACUGCUGGCAGCGGGAACGUUAUAGCACCACCAAUUUCCG ..((((.(((((.....(((((((.(((((.......(...((...((.((((((....)))))).))..))..).......)))))))))))))))))...)))).............. ( -41.44, z-score = -2.05, R) >droEre2.scaffold_4845 7131142 120 - 22589142 UCGUUACUGUUCAAAAUCCGCUGCACAGCAUAAACGGCAUUCCUAUUGGUGGACGCGGGCGUCCAGCAUCGGUGGACACAACUGCUGGCAGCGGCAACGUUAUAGCACCAACAAUUUCUG ..(((.((((.......(((((((.(((((.......(...((...((.((((((....)))))).))..))..).......)))))))))))).......))))....)))........ ( -38.98, z-score = -1.68, R) >droAna3.scaffold_13266 14722586 120 + 19884421 UCGUCACUGUUCAGAAUCCACUGCACAGCCUCAACGGCAUAUCAAUGAGUGGACGCGGGCGUCCAGCGUCGGUGGAUACAACCGCUGGCAGUGGCAACGUCAUGGCGCCAACAAUAACUG ..((((((((.(((.((((((((.((.(((.....)))...........((((((....))))))..)))))))))).......)))))))))))..(((....)))............. ( -42.80, z-score = -1.41, R) >dp4.chr3 5025709 120 + 19779522 UUGUCACCGUUCAGAAUCCGCUGCACAGCCUGAAUGGCCUGCCAAUGGGCGGACGCGGGCGUCCCGCAUCGGUGGAUAAUACAGCGGGCAGCGGCAACGUUAUGGCGCCAGCAAUUACUG ..(((.((((((((.....(((....)))))))))))..((((....)))))))((.((((((..((.(((.((.......)).)))))((((....))))..)))))).))........ ( -46.10, z-score = -0.24, R) >droPer1.super_2 5225077 120 + 9036312 UUGUCACCGUUCAGAAUCCGCUGCACAGCCUGAAUGGCCUGCCAAUGGGCGGACGCGGGCGUCCCGCAUCGGUGGAUAAUACAGCGGGCAGCGGCAACGUUAUGGCGCCGGCAAUUACUG ..(((.((((((((.....(((....)))))))))))..((((....)))))))((.((((((..((.(((.((.......)).)))))((((....))))..)))))).))........ ( -47.90, z-score = -0.41, R) >droWil1.scaffold_180701 950031 120 + 3904529 UUGUCACUGUGCAAAAUCCAUUGCAUAGCCUUAAUGGUCUACCAAUGGGCGGACGCGGGCGUCCAGCUUCGGUGGAUAAUACAACAGUUAGUGGCAAUUUGGUGUCAACAGCUAUUGCGG (((.((((((((((......)))))).((((((((.(((((((...(((((((((....))))).)))).))))))).........))))).))).....)))).)))..((....)).. ( -39.90, z-score = -1.57, R) >droVir3.scaffold_12875 13776058 117 - 20611582 UUGUCGCCGUGCAGAAUCCGCUGCACAGCCUCAAUGGCAUAAGCAUGGGCGGACGCGGGCGUCCGGCUUCUGUAGACAAUAACAAUAGCGCUGGCAAUGGUGCAACCGCGCUUGCCA--- (((((((.((((((......)))))).(((.....)))....))..((.((((((....)))))).))......))))).............((((..(((((....))))))))).--- ( -47.10, z-score = -0.71, R) >droMoj3.scaffold_6496 13396197 105 + 26866924 UUGUUGCCGUUCAGAAUUCGCUGCACAGCCUUAAUGGCAUUAGCUUGGGCGGACGCGGGCGUCCGGCUUCAGUGGACAACAACAAUAGUGCCGGCAACGAUGCAA--------------- ((((((((((.(((......))).)).........((((((((((.((.((((((....)))))).))..)))(.....).....))))))))))))))).....--------------- ( -39.20, z-score = -1.33, R) >droGri2.scaffold_15112 3468758 105 - 5172618 UUGUCACUGUGCAGAAUCCGUUGCAUGGCUUGAAUGGCAUUACGAUGGGCGGACGCGGGCGUCCGGCUUCUGUGGACAACAACAAUAGCGCUGGCAAUGGUGCCA--------------- (((((.....(((((((((((((....(((.....)))....)))))))((((((....))))))..)))))).)))))........(((((......)))))..--------------- ( -36.40, z-score = -0.02, R) >consensus UUGUCACUGUUCAGAAUCCGCUGCACAGCCUAAACGGCAUUCCUAUGGGCGGACGCGGGCGUCCAGCAUCGGUGGACAAAACCGCUGGCAGCGGCAACGUUAUAGCACCACCAAUUUCCG ..((((..........(((((((....((.......)).........(.((((((....)))))).)..))))))).............((((....)))).)))).............. (-23.06 = -22.38 + -0.67)
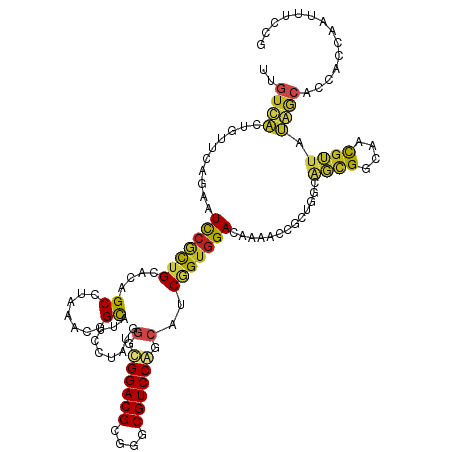
| Location | 10,397,768 – 10,397,862 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.83 |
| Shannon entropy | 0.54500 |
| G+C content | 0.59047 |
| Mean single sequence MFE | -35.39 |
| Consensus MFE | -17.69 |
| Energy contribution | -17.11 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.597597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
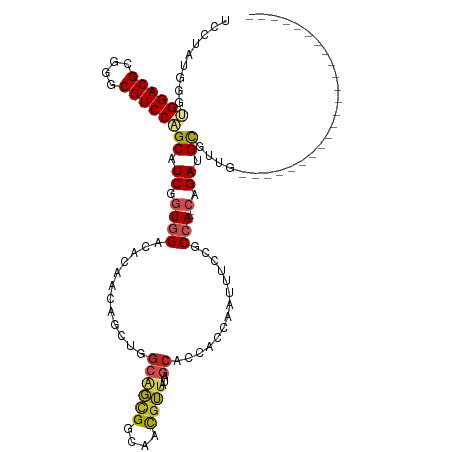
>dm3.chr2R 10397768 94 + 21146708 UCCUAUGGGUGGACGCGGGCGUCCAGCAUCGGUGGACACAACGGCUGGCAGCGGCAACGUUAUAGCACCACCAAUUUCCGCCA---CAGAUGCGUUG----------------------- .((....))((((((....))))))(((((((((((.......((((..((((....)))).))))..........)))))).---..)))))....----------------------- ( -35.53, z-score = -1.14, R) >droSim1.chr2R 9152237 94 + 19596830 UCCUAUGGGUGGACGCGGGCGUCCAGCAUCGGUGGACACAACGGCUGGCAGCGGCAACGUUAUAGCACCACAAAUUUCCGCCA---CAGAUGCGUUG----------------------- .((....))((((((....))))))(((((((((((.......((((..((((....)))).))))..........)))))).---..)))))....----------------------- ( -35.53, z-score = -1.45, R) >droSec1.super_1 7908287 94 + 14215200 UCCUAGGUGUGGACGGGGGCGUCCAGCAUCGGUGGACACAACGGCUGGCAGCGGCAACGUUAUAACACCACAAAUUUCCGCCA---CAGAUGCGUUG----------------------- .........((((((....))))))(((((((((((......((.((..((((....))))....)))).......)))))).---..)))))....----------------------- ( -31.32, z-score = -0.47, R) >droYak2.chr2R 10323413 94 + 21139217 UCCUAUUGGUGGACGCGGGCGUCCAGCAUCGGUGGACACAACUGCUGGCAGCGGGAACGUUAUAGCACCACCAAUUUCCGCCA---CAGAUGCGUUG----------------------- .((....))((((((....))))))(((((((((((......(((((..((((....)))).))))).........)))))).---..)))))....----------------------- ( -35.16, z-score = -1.48, R) >droEre2.scaffold_4845 7131182 94 - 22589142 UCCUAUUGGUGGACGCGGGCGUCCAGCAUCGGUGGACACAACUGCUGGCAGCGGCAACGUUAUAGCACCAACAAUUUCUGCCA---CAGAUGCGUUG----------------------- .((....))((((((....))))))(((((.((((.......(((((..((((....)))).)))))....((.....)))))---).)))))....----------------------- ( -34.20, z-score = -1.54, R) >droAna3.scaffold_13266 14722626 94 + 19884421 AUCAAUGAGUGGACGCGGGCGUCCAGCGUCGGUGGAUACAACCGCUGGCAGUGGCAACGUCAUGGCGCCAACAAUAACUGCCA---CAGAUGCUAUG----------------------- ..........(((((....)))))(((((((((((......)))))(((((((....)((....))..........)))))).---..))))))...----------------------- ( -35.10, z-score = -1.17, R) >dp4.chr3 5025749 94 + 19779522 GCCAAUGGGCGGACGCGGGCGUCCCGCAUCGGUGGAUAAUACAGCGGGCAGCGGCAACGUUAUGGCGCCAGCAAUUACUGCCA---CCGAGGUCCUG----------------------- (((....)))(((((((((...))))).(((((((........((.(((((((....)))).....))).))........)))---)))).))))..----------------------- ( -43.79, z-score = -2.09, R) >droPer1.super_2 5225117 94 + 9036312 GCCAAUGGGCGGACGCGGGCGUCCCGCAUCGGUGGAUAAUACAGCGGGCAGCGGCAACGUUAUGGCGCCGGCAAUUACUGCCA---CCGAGGUUCUG----------------------- (((....)))(((((((((...))))).(((((((........((.(((((((....)))).....))).))........)))---)))).))))..----------------------- ( -42.69, z-score = -1.68, R) >droWil1.scaffold_180701 950071 120 + 3904529 ACCAAUGGGCGGACGCGGGCGUCCAGCUUCGGUGGAUAAUACAACAGUUAGUGGCAAUUUGGUGUCAACAGCUAUUGCGGCCAGUACUGAUGCAGGGCAAAUGCAUCAUACUCACAAUAA .(((..(((((((((....))))).))))...)))...............((((((((..(.((....)).).)))))....((((.(((((((.......))))))))))))))..... ( -36.90, z-score = -0.77, R) >droVir3.scaffold_12875 13776098 91 - 20611582 AAGCAUGGGCGGACGCGGGCGUCCGGCUUCUGUAGACAAUAACAAUAGCGCUGGCAAUGGU---GCAACCGCGCUUGCCACCA---ACGAUGCGCUG----------------------- ..((((.(.((((((....))))))(((..(((........)))..)))(.(((((..(((---((....)))))))))).).---.).))))....----------------------- ( -35.20, z-score = -0.75, R) >droGri2.scaffold_15112 3468798 79 - 5172618 UACGAUGGGCGGACGCGGGCGUCCGGCUUCUGUGGACAACAACAAUAGCGCUGGCAAUGGU---GC------------CACAA---AUGAUGCACUG----------------------- ((((..((.((((((....)))))).))..))))...........(((.(((((((....)---))------------))...---.....)).)))----------------------- ( -23.90, z-score = -0.05, R) >consensus UCCUAUGGGUGGACGCGGGCGUCCAGCAUCGGUGGACACAACAGCUGGCAGCGGCAACGUUAUAGCACCACCAAUUUCCGCCA___CAGAUGCGUUG_______________________ ........(((((((....))))).))...(((((............((((((....))))...))...........)))))...................................... (-17.69 = -17.11 + -0.58)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:22:43 2011