
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,334,817 – 10,334,915 |
| Length | 98 |
| Max. P | 0.686358 |

| Location | 10,334,817 – 10,334,915 |
|---|---|
| Length | 98 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 76.98 |
| Shannon entropy | 0.46974 |
| G+C content | 0.48144 |
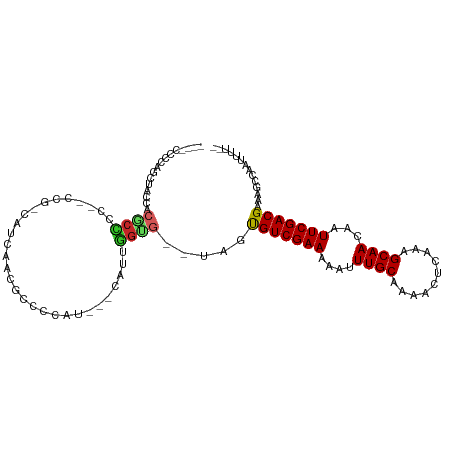
| Mean single sequence MFE | -19.51 |
| Consensus MFE | -10.07 |
| Energy contribution | -10.16 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.686358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
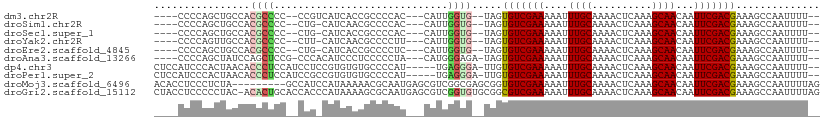
>dm3.chr2R 10334817 98 + 21146708 ----CCCCAGCUGCCACGCCCC--CCGUCAUCACCGCCCCAC---CAUUGGUG--UAGUGUCGAAAAAUUUGCAAAACUCAAAGCAACAAUUCGACGAAAGCCAAUUUU-- ----.....(((..(.......--...........((..(((---(...))))--..))((((((....((((..........))))...)))))))..))).......-- ( -17.30, z-score = -1.30, R) >droSim1.chr2R 9099500 97 + 19596830 ----CCCCAGCUGCCACGCCCC--CUG-CAUCAACGCCCCAC---CAUUGGUG--UAGUGUCGAAAAAUUUGCAAAACUCAAAGCAACAAUUCGACGAAAGCCAAUUUU-- ----.....(((..(.......--(((-((((((.(......---).))))))--))).((((((....((((..........))))...)))))))..))).......-- ( -21.10, z-score = -2.32, R) >droSec1.super_1 7856465 97 + 14215200 ----CCCCAGCUGCCACGCCCC--CUG-CAUCACCGCCCCAC---CAUUGGUG--UAGUGUCGAAAAAUUUGCAAAACUCAAAGCAACAAUUCGACGAAAGCCAAUUUU-- ----.....(((..(.......--(((-(((((.........---...)))))--))).((((((....((((..........))))...)))))))..))).......-- ( -19.50, z-score = -1.80, R) >droYak2.chr2R 10269231 97 + 21139217 ----CCCCAGUUGCCACGCCCC--CUU-CAUCAACGCCCCUU---CAUUGGUG--UAGUGUCGAAAAAUUUGCAAAACUCAAAGCAACAAUUCGACGAAAGCCAAUUUU-- ----.....((((.........--...-...)))).......---.((((((.--...(((((((....((((..........))))...)))))))...))))))...-- ( -17.56, z-score = -1.62, R) >droEre2.scaffold_4845 7078782 97 - 22589142 ----CCCCAGCUGCCACGCCCC--CUG-CAUCACCGCCCCUC---CAUUGGUG--UAGUGUCGAAAAAUUUGCAAAACUCAAAGCAACAAUUCGACGAAAGCCAAUUUU-- ----.....(((..(.......--(((-(((((.........---...)))))--))).((((((....((((..........))))...)))))))..))).......-- ( -19.50, z-score = -1.87, R) >droAna3.scaffold_13266 14673359 100 + 19884421 ----CCCCAGCUAUCCAGCUCCG-CCCACAUCCCUCCCCCUA---CAUGGGAGA-UAGUGUCGAAAAAUUUGCAAAACUCAAAGCAACAAUUCGACGAAAGCCAAUUUU-- ----.....(((.((........-.........(((((....---...))))).-....((((((....((((..........))))...)))))))).))).......-- ( -20.40, z-score = -2.78, R) >dp4.chr3 15431851 103 - 19779522 CUCCAUCCCACUAACACCCUCCAUCCUCCGUGUGUGCCCCAU-----UGAGGGA-UUGUGUCGAAAAAUUUGCAAAACUCAAAGCAACAAUUCGACGAAAGCCAAUUUU-- (((.((..(((..((((............)))))))....))-----.)))((.-...(((((((....((((..........))))...)))))))....))......-- ( -17.00, z-score = -0.44, R) >droPer1.super_2 4499779 103 - 9036312 CUCCAUCCCACUAACACCCUCCAUCCGCCGUGUGUGCCCCAU-----UGAGGGA-UUGUGUCGAAAAAUUUGCAAAACUCAAAGCAACAAUUCGACGAAAGCCAAUUUU-- ................(((((.((..(((....).))...))-----.))))).-...(((((((....((((..........))))...)))))))............-- ( -17.60, z-score = -0.31, R) >droMoj3.scaffold_6496 13324963 102 + 26866924 ACACCUCCCUCUA---------GCCAUCCAUAAAAACGCAAUGAGCGUCGGCGAGCGGUGUCGAAAAAUUUGCAAAACUCAAAGCAACAAUUCGACGAAAGCCAAUUUUAG .............---------........(((((((((.....)))).(((......(((((((....((((..........))))...)))))))...)))..))))). ( -19.70, z-score = -0.69, R) >droGri2.scaffold_15112 3402260 110 - 5172618 CUACCUCCCCCUAC-ACACUGCACCACCCAUAAAAGCGCAAUGAGCGUCGGUGUGCGGCGUCGAAAAAUUUGCAAAACUCAAAGCAACAAUUCGACGAAAGCCAAUUUUAG ..............-...((((((.(((.......((((.....)))).)))))))))(((((((....((((..........))))...))))))).............. ( -25.40, z-score = -2.15, R) >consensus ____CCCCAGCUACCACGCCCC__CCG_CAUCAACGCCCCAU___CAUUGGUG__UAGUGUCGAAAAAUUUGCAAAACUCAAAGCAACAAUUCGACGAAAGCCAAUUUU__ .........((((...((((.............................))))..))))((((((....((((..........))))...))))))............... (-10.07 = -10.16 + 0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:22:35 2011