
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,316,186 – 10,316,294 |
| Length | 108 |
| Max. P | 0.721245 |

| Location | 10,316,186 – 10,316,294 |
|---|---|
| Length | 108 |
| Sequences | 13 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 68.74 |
| Shannon entropy | 0.71142 |
| G+C content | 0.46561 |
| Mean single sequence MFE | -20.39 |
| Consensus MFE | -8.71 |
| Energy contribution | -8.85 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.18 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.721245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10316186 108 - 21146708 -UCUACUACCCGUACUCAUCUAUUCCCUAUAUAUCUCUACUUUCAGCCCCUGAACGAGGACAAAUUCCUCUGCUUUAGCUGCAACAACUGGCUGAUCAACUGGCACUCC -.........................................((((((.......(((((.....)))))(((.......)))......)))))).............. ( -18.10, z-score = -1.71, R) >droSim1.chr2R 9080058 108 - 19596830 -UAGACAACCCGUACUCAUCUAUGCUCUAUAUGUCUCUACUUUCAGCCAUUGAAUGAGGACAAAUUCCUCUGCUUCAGCUGCAACAACUGGCUGAUCAACUGGCACUCC -((((.....((((......)))).))))..((((.......((((((((((...(((((.....)))))(((.......))).))).)))))))......)))).... ( -24.62, z-score = -1.98, R) >droSec1.super_1 7838007 108 - 14215200 -UAGGCAACCCGUACUCAUCUAUGCCCUAUAUGUCUCUACUUUCAGCCAUUGAAUGAGGACAAAUUCCUCUGCUUCAGCUGCAACAACUGGCUGAUCAACUGGCACUCC -..((((...............)))).....((((.......((((((((((...(((((.....)))))(((.......))).))).)))))))......)))).... ( -27.68, z-score = -2.43, R) >droYak2.chr2R 10249323 106 - 21139217 -UGAACAUCCUGUACUCAUCUAUGCCUUGUA--UCUGCACUUUCAGCCCCUGAAUGAGGACAAGUUCCUCUGCUUCAGCUGCAACAACUGGCUGAUCAACUGGCACUCC -.....................(((((((.(--((.((.......((..(((((.(((((.....)))))...)))))..))........)).))))))..)))).... ( -23.36, z-score = -0.33, R) >droEre2.scaffold_4845 7060104 106 + 22589142 -UGGACAUCCCGUGCUCAUCUAUUCCUUGUC--UCGCUACUUUCAGCCCCUCAAUGAGGACAAGUUCCUCUGCUUCAGCUGCAACAACUGGCUGAUCAACUGGCACUCC -.(((((....(((......)))....))))--).((((...((((((.......(((((.....)))))(((.......)))......)))))).....))))..... ( -24.80, z-score = -0.81, R) >droAna3.scaffold_13266 14657370 102 - 19884421 -----AUCCCCAGACUAACCUAUUCUCUUCU--CUUAUCCUUACAGCCUUUGAACGAGGACAAAUUCCUAUGCUUCAGCUGCAACAACUGGCUCAUCAACUGGCAUUCC -----....((((..................--..((....)).((((.(((....((((.....)))).(((.......))).)))..))))......))))...... ( -14.00, z-score = -0.01, R) >dp4.chr3 15413693 92 + 19779522 ---------------UCACCCGCAUUCUUCU--UCUCCAAUUACAGCCCCUCAAUGAGGAUAAGUUUCUGUGCUUCAGCUGCAACAACUGGCUUAUCAAUUGGCACUCC ---------------................--...((((((..((((((((...))))...((((..(((((....)).)))..))))))))....))))))...... ( -17.70, z-score = -0.80, R) >droPer1.super_2 4482502 90 + 9036312 -----------------ACCCGCAUUCUUCU--UCUCCAAUUACAGCCCCUCAAUGAGGAUAAGUUUCUGUGCUUCAGCUGCAACAACUGGCUUAUCAAUUGGCACUCC -----------------..............--...((((((..((((((((...))))...((((..(((((....)).)))..))))))))....))))))...... ( -17.70, z-score = -0.81, R) >droWil1.scaffold_181009 136976 100 - 3585778 --------ACCACACAUCUCUUCUUUUUUUAUUUUUCU-CUUGUAGCCCUUGAAUGAGGAUAAAUUUCUAUGCUUUAGCUGCAACAAUUGGCUAAUCAAUUGGCAUUCC --------..............................-.(((((((((((....))))(((......)))......)))))))(((((((....)))))))....... ( -14.70, z-score = -0.16, R) >droMoj3.scaffold_6496 13303025 99 - 26866924 ---------AACAACUCUCUCCCUCUCUUUCUCUCUGU-CUUGCAGCCCUUGAAUGAGGAUAAAUUCCUGUGCUUCAGCUGCAACAAUUGGCUAAUCAAUUGGCACUCG ---------..........................(((-.(((((((...((((..((((.....))))....)))))))))))(((((((....)))))))))).... ( -19.70, z-score = -1.38, R) >droVir3.scaffold_12875 13688817 107 + 20611582 UCUGUCUCUCUUGGUCUUCUCUCUCUCUCUCUGUCU-C-CUUGCAGCCUCUGAAUGAGGAUAAAUUCCUGUGCUUCAGCUGCAACAAUUGGCUAAUCAACUGGCACUCG ..((((....((((((..........(.....)...-.-.(((((((...((((..((((.....))))....))))))))))).....))))))......)))).... ( -19.10, z-score = -0.21, R) >droGri2.scaffold_15112 3378944 83 + 5172618 ------------------------CUCUCUGUGUCU-G-CUUACAGCCGCUGAAUGAGGAUAAAUUCCUGUGCUUUAGCUGCAACAAUUGGCUCAUAAACUGGCACUCG ------------------------......(((((.-(-.(((.((((........((((.....)))).(((.......)))......))))..))).).)))))... ( -18.40, z-score = -0.29, R) >anoGam1.chr3L 15621814 108 - 41284009 -CCUCCACUAACCUUUUCAACAUUCCCUACUCUGCAGGCCGAGCGGGGUGAUAACGCGGACAAACACAUCUGCUUCAGCUGCAACAACUGGCUGAUCAAUUGGUACUCG -.........(((...........(((..(((.(....).)))..)))(((....(((((........))))).((((((.........)))))))))...)))..... ( -25.20, z-score = -0.79, R) >consensus _____C__CCCGUACUCAUCUAUUCUCUUUAU_UCUCUACUUUCAGCCCCUGAAUGAGGACAAAUUCCUCUGCUUCAGCUGCAACAACUGGCUGAUCAACUGGCACUCC ............................................((((........((((.....)))).(((.......)))......))))................ ( -8.71 = -8.85 + 0.14)

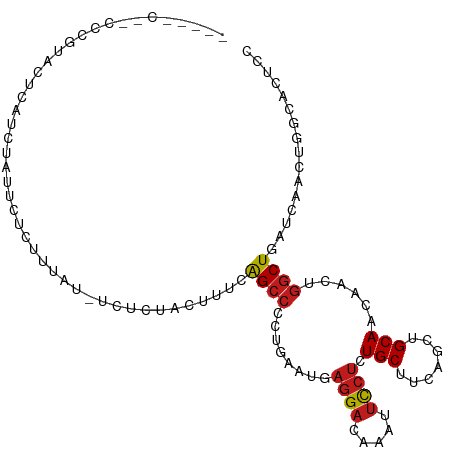
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:22:30 2011