
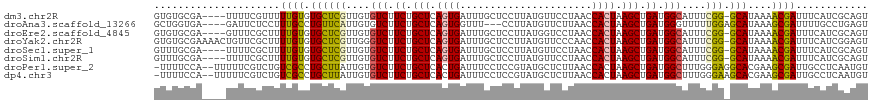
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,264,215 – 10,264,329 |
| Length | 114 |
| Max. P | 0.670975 |

| Location | 10,264,215 – 10,264,329 |
|---|---|
| Length | 114 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.44 |
| Shannon entropy | 0.36198 |
| G+C content | 0.45084 |
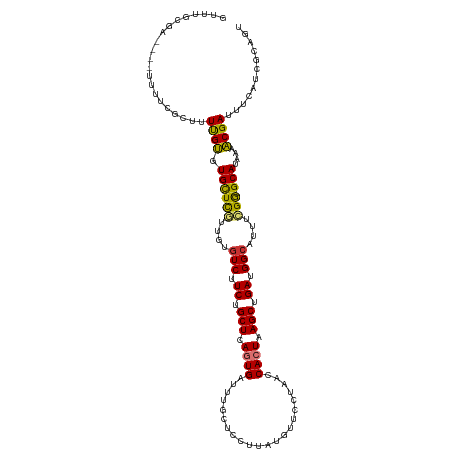
| Mean single sequence MFE | -32.55 |
| Consensus MFE | -16.03 |
| Energy contribution | -15.36 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.670975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
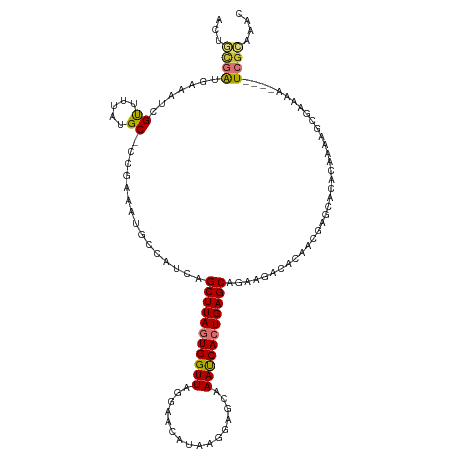
>dm3.chr2R 10264215 114 + 21146708 GUGUGCGA----UUUUCGUUUUUGUGUGCUCGUUGUGUCUUCUGCUCAGUGAUUUGCUCCUUAUGUUCCUAACCACUAAGCUGAUGGCAUUUCGG-GCAUAAAACGAUUUCAUCGCAGU ...(((((----(..((((((...((((((((..(((((.((.(((.((((.((.((.......))....)).)))).))).)).)))))..)))-)))))))))))....)))))).. ( -40.10, z-score = -4.96, R) >droAna3.scaffold_13266 14610293 112 + 19884421 GCUGGUGA----GAUUCUCCUUUGCCUGUUCAUUGUGUCUUCUGCUCAGUGGUUU---CCUUAUGUUCUUAACCACUAAGCUGAUGGGUUUUUGGAGCAUAAAGCGAUUUUGCCUGAGU ...((..(----((((...(((((..(((((...(..(((((.(((.(((((((.---............))))))).))).)).)))..)...)))))))))).)))))..))..... ( -29.02, z-score = -0.80, R) >droEre2.scaffold_4845 7012194 114 - 22589142 GUGUGCGA----GUUUCGCUUUUGUGUGCUCGUUGUGUCUUCUGCUCAGUGAUUUGCUCCUUAUGGUCCUAACCACUAAGCUGAUGGCAUUUCGG-GCAUAAAACGAUUUCAUCGCAGU ...(((((----...(((......((((((((..(((((.((.(((.((((.((.(..((....))..).)).)))).))).)).)))))..)))-)))))...))).....))))).. ( -39.60, z-score = -3.77, R) >droYak2.chr2R 10201833 118 + 21139217 GUGUGCGAAAACUGUUCGCUUUUGUGUGCUCGUUGGGUCUUCUGCUCAGUGAUUUGCUCCUUAUGUUCCCAACCACUAAGCUGAUGGCAUUUCGG-GCAUAAAACGAUUUCAUCGGAGU ....(((((.....))))).....((((((((....(((.((.(((.((((....((.......)).......)))).))).)).)))....)))-)))))...((((...)))).... ( -30.00, z-score = -0.25, R) >droSec1.super_1 7773438 114 + 14215200 GUUUGCGA----UUUUCGCUUUUGUGUGCUCGUUGUGUCUUCUGCUCAGUGAUUUGCUCCUUAUGUUCCUAACCACUAAGCUGAUGGCAUUUCGG-GCAUAAAACGAUUUCAUCGCAGU ..((((((----(..(((......((((((((..(((((.((.(((.((((.((.((.......))....)).)))).))).)).)))))..)))-)))))...)))....))))))). ( -37.20, z-score = -4.04, R) >droSim1.chr2R 9028069 114 + 19596830 GUUUGCGA----UUUUCGCUUUUGUGUGCUCGUUGUGUCUUCUGCUCAGUGAUUUGCUCCUUAUGUUCCUAACCACUAAGCUGAUGGCAUUUCGG-GCAUAAAACGAUUUCAUCGCAGU ..((((((----(..(((......((((((((..(((((.((.(((.((((.((.((.......))....)).)))).))).)).)))))..)))-)))))...)))....))))))). ( -37.20, z-score = -4.04, R) >droPer1.super_2 4436775 116 - 9036312 -UUUUCCA--UUUUUCGUCUGUCGCCUGCUUAUUGUGUCUUCUGCUCACUGAUUUCCUCCGUAUGCUCUUAACCACUAAGCUGAUGGCUUUGGGAGGCACGAAGCGAUUGCCUCAAUGU -.....((--((......(((..(((.(((((.((.((.....((..((.((.....)).))..)).....)))).)))))....)))..)))((((((.........)))))))))). ( -24.50, z-score = 0.12, R) >dp4.chr3 15367765 116 - 19779522 -UUUUCCA--UUUUUCGUCUGUCGCCUGCUUAUUGUGUCUUCUGCUCACUGAUUUCCUCCGUAUGCUCUUAACCACUAAGCUGAUGGCUUUGGGAAGCACGAAGCGAUUGCCUCAAUGU -.......--.....(((.((..((.((((..(((((.((((.((..((.((.....)).))..))......(((..((((.....))))))))))))))))))))...))..))))). ( -22.80, z-score = 0.21, R) >consensus GUUUGCGA____UUUUCGCUUUUGUGUGCUCGUUGUGUCUUCUGCUCAGUGAUUUGCUCCUUAUGUUCCUAACCACUAAGCUGAUGGCAUUUCGG_GCAUAAAACGAUUUCAUCGCAGU .....................((((.((((((....(((.((.(((.((((......................)))).))).)).)))....))).)))....))))............ (-16.03 = -15.36 + -0.67)
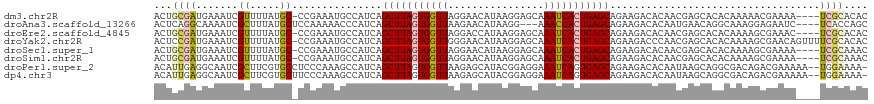
| Location | 10,264,215 – 10,264,329 |
|---|---|
| Length | 114 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.44 |
| Shannon entropy | 0.36198 |
| G+C content | 0.45084 |
| Mean single sequence MFE | -28.93 |
| Consensus MFE | -13.57 |
| Energy contribution | -13.59 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.517247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10264215 114 - 21146708 ACUGCGAUGAAAUCGUUUUAUGC-CCGAAAUGCCAUCAGCUUAGUGGUUAGGAACAUAAGGAGCAAAUCACUGAGCAGAAGACACAACGAGCACACAAAAACGAAAA----UCGCACAC ..((((((....(((((((.(((-.((...((.(.((.(((((((((((.(...(....)...).))))))))))).)).).))...)).)))....)))))))..)----)))))... ( -31.90, z-score = -3.81, R) >droAna3.scaffold_13266 14610293 112 - 19884421 ACUCAGGCAAAAUCGCUUUAUGCUCCAAAAACCCAUCAGCUUAGUGGUUAAGAACAUAAGG---AAACCACUGAGCAGAAGACACAAUGAACAGGCAAAGGAGAAUC----UCACCAGC .(((..((....((((.....))...............(((((((((((............---.)))))))))))............))....))....)))....----........ ( -23.82, z-score = -1.12, R) >droEre2.scaffold_4845 7012194 114 + 22589142 ACUGCGAUGAAAUCGUUUUAUGC-CCGAAAUGCCAUCAGCUUAGUGGUUAGGACCAUAAGGAGCAAAUCACUGAGCAGAAGACACAACGAGCACACAAAAGCGAAAC----UCGCACAC ..(((((.(...(((((((.(((-.((...((.(.((.(((((((((((.(..((....))..).))))))))))).)).).))...)).)))....)))))))..)----)))))... ( -32.50, z-score = -3.30, R) >droYak2.chr2R 10201833 118 - 21139217 ACUCCGAUGAAAUCGUUUUAUGC-CCGAAAUGCCAUCAGCUUAGUGGUUGGGAACAUAAGGAGCAAAUCACUGAGCAGAAGACCCAACGAGCACACAAAAGCGAACAGUUUUCGCACAC .(((.((((....((((((....-..)))))).)))).(((((((((((.....(....).....)))))))))))............))).........(((((.....))))).... ( -27.40, z-score = -1.01, R) >droSec1.super_1 7773438 114 - 14215200 ACUGCGAUGAAAUCGUUUUAUGC-CCGAAAUGCCAUCAGCUUAGUGGUUAGGAACAUAAGGAGCAAAUCACUGAGCAGAAGACACAACGAGCACACAAAAGCGAAAA----UCGCAAAC ..((((((....(((((((.(((-.((...((.(.((.(((((((((((.(...(....)...).))))))))))).)).).))...)).)))....)))))))..)----)))))... ( -32.20, z-score = -3.53, R) >droSim1.chr2R 9028069 114 - 19596830 ACUGCGAUGAAAUCGUUUUAUGC-CCGAAAUGCCAUCAGCUUAGUGGUUAGGAACAUAAGGAGCAAAUCACUGAGCAGAAGACACAACGAGCACACAAAAGCGAAAA----UCGCAAAC ..((((((....(((((((.(((-.((...((.(.((.(((((((((((.(...(....)...).))))))))))).)).).))...)).)))....)))))))..)----)))))... ( -32.20, z-score = -3.53, R) >droPer1.super_2 4436775 116 + 9036312 ACAUUGAGGCAAUCGCUUCGUGCCUCCCAAAGCCAUCAGCUUAGUGGUUAAGAGCAUACGGAGGAAAUCAGUGAGCAGAAGACACAAUAAGCAGGCGACAGACGAAAAA--UGGAAAA- .((((((((((.........)))))).....(((....(((((.(((((....((.(((.((.....)).))).))....))).)).))))).)))...........))--)).....- ( -26.20, z-score = -0.39, R) >dp4.chr3 15367765 116 + 19779522 ACAUUGAGGCAAUCGCUUCGUGCUUCCCAAAGCCAUCAGCUUAGUGGUUAAGAGCAUACGGAGGAAAUCAGUGAGCAGAAGACACAAUAAGCAGGCGACAGACGAAAAA--UGGAAAA- .(((((((((....)))))(((((((....((((((.......))))))....((.(((.((.....)).))).)).)))).)))......................))--)).....- ( -25.20, z-score = -0.16, R) >consensus ACUGCGAUGAAAUCGUUUUAUGC_CCGAAAUGCCAUCAGCUUAGUGGUUAGGAACAUAAGGAGCAAAUCACUGAGCAGAAGACACAACGAGCACACAAAAGCGAAAA____UCGCAAAC ...((((.......((.....))...............(((((((((((................)))))))))))...................................)))).... (-13.57 = -13.59 + 0.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:22:29 2011