
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,250,521 – 10,250,579 |
| Length | 58 |
| Max. P | 0.982117 |

| Location | 10,250,521 – 10,250,579 |
|---|---|
| Length | 58 |
| Sequences | 7 |
| Columns | 69 |
| Reading direction | forward |
| Mean pairwise identity | 74.07 |
| Shannon entropy | 0.46660 |
| G+C content | 0.46314 |
| Mean single sequence MFE | -17.37 |
| Consensus MFE | -8.31 |
| Energy contribution | -9.66 |
| Covariance contribution | 1.35 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.982117 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
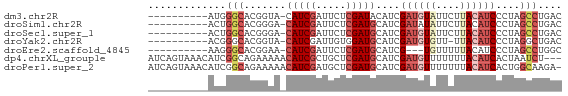
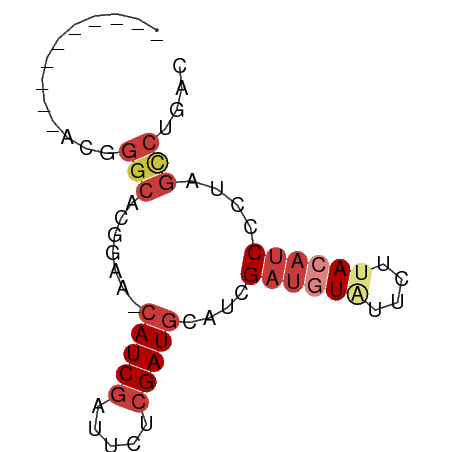
>dm3.chr2R 10250521 58 + 21146708 ----------AUGGGCACGGUA-CAUCGAUUCUCGAUACAUCGAUGUAUUCUUACAUCCCUAGCCUGAC ----------..((((..(((.-.((((.....))))..)))((((((....))))))....))))... ( -16.70, z-score = -2.56, R) >droSim1.chr2R 9014853 58 + 19596830 ----------ACUGGCACGGGA-CAUCGAUUCUCGAUGCAUCGAUAUAUUCUUACAUCCCUAGCCUGAC ----------...(((..((((-(((((.....)))))....((.....)).....))))..))).... ( -15.50, z-score = -2.47, R) >droSec1.super_1 7758232 58 + 14215200 ----------ACUGGCACGGGA-CAUCGAUUCUCGAUGCAUCGAUGUAUUCUUACAUCCCUAGCCUGAC ----------...(((..((..-(((((.....)))))..))((((((....))))))....))).... ( -18.30, z-score = -2.96, R) >droYak2.chr2R 10187943 57 + 21139217 ----------ACGGGCACGGUA-CAUCGAUUGUGGAUGCAUCGAUGUGUU-UUACAUCCCUAGGCUGAC ----------.(((.(..((((-(((((((.((....)))))))))))..-.......))..).))).. ( -13.90, z-score = -0.03, R) >droEre2.scaffold_4845 6998384 55 - 22589142 ----------AAGGGCACGGAA-CAUCGAUUCUCGAUGCAUCG---UGUUUUUACAUCCCUAGCCUGGC ----------((((((((((..-(((((.....)))))..)))---))))))).....((......)). ( -16.60, z-score = -1.98, R) >dp4.chrXL_group1e 6499613 66 + 12523060 AUCAGUAAACAUCGGCAGAAAAACAUCGCUGCUCGAUGCAUCGAUGUUUUUUUACAUCACUAAUCU--- ..............(.((((((((((((.(((.....))).)))))))))))).)...........--- ( -17.10, z-score = -3.60, R) >droPer1.super_2 3166322 68 + 9036312 AUCAGUAAACAUCGGCAGAAAAACAUCGAUGCUCGAUGCAUCGAUGUUUUUUUACAUCACUGGCAAGA- .(((((........(.((((((((((((((((.....)))))))))))))))).)...))))).....- ( -23.50, z-score = -4.62, R) >consensus __________ACGGGCACGGAA_CAUCGAUUCUCGAUGCAUCGAUGUAUUCUUACAUCCCUAGCCUGAC .............(((.......(((((.....)))))....((((((....))))))....))).... ( -8.31 = -9.66 + 1.35)
| Location | 10,250,521 – 10,250,579 |
|---|---|
| Length | 58 |
| Sequences | 7 |
| Columns | 69 |
| Reading direction | reverse |
| Mean pairwise identity | 74.07 |
| Shannon entropy | 0.46660 |
| G+C content | 0.46314 |
| Mean single sequence MFE | -18.71 |
| Consensus MFE | -9.20 |
| Energy contribution | -9.94 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.981782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
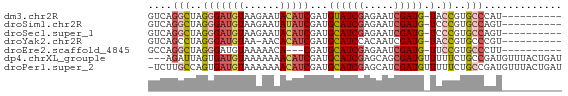
>dm3.chr2R 10250521 58 - 21146708 GUCAGGCUAGGGAUGUAAGAAUACAUCGAUGUAUCGAGAAUCGAUG-UACCGUGCCCAU---------- ....(((..((((((((....))))))....(((((.....)))))-..))..)))...---------- ( -16.10, z-score = -1.57, R) >droSim1.chr2R 9014853 58 - 19596830 GUCAGGCUAGGGAUGUAAGAAUAUAUCGAUGCAUCGAGAAUCGAUG-UCCCGUGCCAGU---------- ....(((..((((((((....))))))...((((((.....)))))-).))..)))...---------- ( -18.00, z-score = -2.17, R) >droSec1.super_1 7758232 58 - 14215200 GUCAGGCUAGGGAUGUAAGAAUACAUCGAUGCAUCGAGAAUCGAUG-UCCCGUGCCAGU---------- ....(((..((((((((....))))))...((((((.....)))))-).))..)))...---------- ( -19.90, z-score = -2.66, R) >droYak2.chr2R 10187943 57 - 21139217 GUCAGCCUAGGGAUGUAA-AACACAUCGAUGCAUCCACAAUCGAUG-UACCGUGCCCGU---------- .........((((((...-...((((((((.........)))))))-)..))).)))..---------- ( -13.60, z-score = -0.90, R) >droEre2.scaffold_4845 6998384 55 + 22589142 GCCAGGCUAGGGAUGUAAAAACA---CGAUGCAUCGAGAAUCGAUG-UUCCGUGCCCUU---------- ........((((.........((---((..((((((.....)))))-)..)))))))).---------- ( -16.40, z-score = -1.78, R) >dp4.chrXL_group1e 6499613 66 - 12523060 ---AGAUUAGUGAUGUAAAAAAACAUCGAUGCAUCGAGCAGCGAUGUUUUUCUGCCGAUGUUUACUGAU ---..((((((((.(((.((((((((((.(((.....))).)))))))))).)))......)))))))) ( -21.10, z-score = -3.54, R) >droPer1.super_2 3166322 68 - 9036312 -UCUUGCCAGUGAUGUAAAAAAACAUCGAUGCAUCGAGCAUCGAUGUUUUUCUGCCGAUGUUUACUGAU -......((((((.(((.((((((((((((((.....)))))))))))))).)))......)))))).. ( -25.90, z-score = -5.25, R) >consensus GUCAGGCUAGGGAUGUAAAAAUACAUCGAUGCAUCGAGAAUCGAUG_UUCCGUGCCCAU__________ ....(((..(((((((......)))))....(((((.....)))))...))..)))............. ( -9.20 = -9.94 + 0.74)
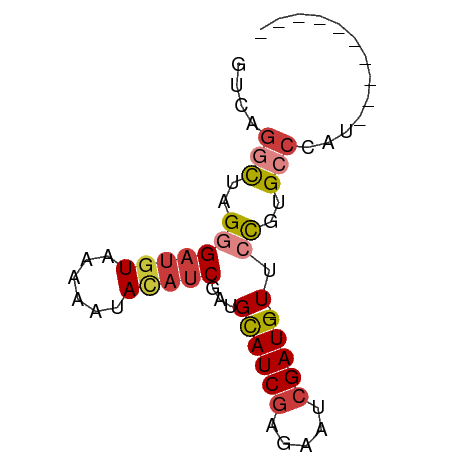
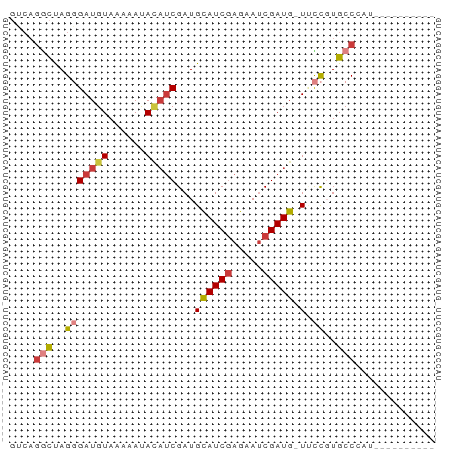
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:22:27 2011