| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,248,056 – 10,248,160 |
| Length | 104 |
| Max. P | 0.993458 |

| Location | 10,248,056 – 10,248,160 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 59.67 |
| Shannon entropy | 0.78187 |
| G+C content | 0.48470 |
| Mean single sequence MFE | -31.02 |
| Consensus MFE | -6.58 |
| Energy contribution | -5.35 |
| Covariance contribution | -1.23 |
| Combinations/Pair | 2.12 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.21 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.855527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
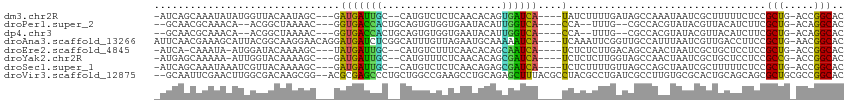
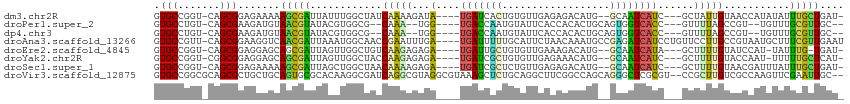
>dm3.chr2R 10248056 104 + 21146708 -AUCAGCAAAUAUAUGGUUACAAUAGC---GAUGAUUGC--CAUGUCUCUCAACACAGUGAUCA----UAUCUUUUGAUAGCCAAAUAAUCGCUUUUUCUCCGCUG-ACCGGCAC -...(((...(((.(((((((((.((.---.(((((..(--..(((......)))..)..))))----)..)).))).)))))).)))...)))........(((.-...))).. ( -24.40, z-score = -1.98, R) >droPer1.super_2 4614440 99 + 9036312 --GCAACGCAAACA--ACGGCUAAAAC---GGUGACCACUGCAGUGUGGUGAAUACAUUGGUCA----CCA--UUUG--CGCCACGUAUACGUUACAUCUUCGCUG-ACAGGCAC --((((.((.....--...))......---((((((((.....(((((.....)))))))))))----)).--.)))--)(((..(((.....))).((......)-)..))).. ( -27.30, z-score = -0.41, R) >dp4.chr3 4426673 99 + 19779522 --GCAACGCAAACA--ACGGCUAAAAC---GGUGACCACUGCAGUGUGGUGAAUACAUUGGUCA----CCA--UUUG--CGCCACGUAUACGUUACAUCUUCGCUG-ACAGGCAC --((((.((.....--...))......---((((((((.....(((((.....)))))))))))----)).--.)))--)(((..(((.....))).((......)-)..))).. ( -27.30, z-score = -0.41, R) >droAna3.scaffold_13266 14593597 110 + 19884421 AUUCAACGAAAGCAUUACGGCAAGGAACAGGAUGAUCUCGGCAUUUGUUAGAAUGCAAAAAUCA----UCAAAUUCGGUUGCCAUUUAAUCGUUGACCUUCCGCUG-AACGGCAC ..(((((((..(.....)(((((.(((...((((((....((((((....))))))....))))----))...)))..)))))......)))))))......(((.-...))).. ( -30.70, z-score = -2.33, R) >droEre2.scaffold_4845 6994258 102 - 22589142 -AUCA-CAAAUA-AUGGAUACAAAAGC---UAUGAUUGC--CAUGUCUUUCAACACAGCAAUCA----UCUCUCUUGACAGCCAACUAAUCGCUGCUCCUCCGCUG-ACCGGCAC -....-......-..(((..(((.((.---.((((((((--..(((......)))..)))))))----)..)).))).((((.........))))....)))(((.-...))).. ( -23.00, z-score = -2.64, R) >droYak2.chr2R 10183932 103 + 21139217 -AUGAGCAAAAA-AUUGGUACAAAAGC---GAUGAUUGC--CAUGUUUCUCAACACAGCGAUCA----UCUCUCUUGGUAGCCAACUAAUCGCUGCUCCUCCGCCG-ACCGGCAC -..(((((....-.(((((.(((.((.---(((((((((--..((((....))))..)))))))----)).)).)))...)))))........)))))....(((.-...))).. ( -33.32, z-score = -3.95, R) >droSec1.super_1 7754230 104 + 14215200 -AUCAGCAAAUAAAUCGUUACAAAAGC---GAUGAUUGC--CAUGUCUCUCAACAGAGCGAUCA----UCUCUUUUGUUAGCCAGCUAAUCGCUUUUUCUCCGCUG-ACCGGCAC -.(((((.........((((((((((.---(((((((((--..(((......)))..)))))))----)).)))))).)))).(((.....)))........))))-)....... ( -33.30, z-score = -4.55, R) >droVir3.scaffold_12875 13551853 111 + 20611582 --GCAAUUCGAACUUGGCGACAAGCGG--ACGCGAGCCCUGCUGGCCGAAGCCUGCAGAGCUUUACGCCUACGCCUGAUCGCCUUGUGCGCACUGCAGCAGCGCUGCGCCGGCAC --((.......((..(((((((.(((.--..((((((.((((.(((....))).)))).)))...)))...))).)).)))))..))(((((.(((....))).)))))..)).. ( -48.80, z-score = -1.57, R) >consensus _AUCAACAAAAACAUGGCGACAAAAGC___GAUGAUCGC__CAUGUCUCUCAACACAGCGAUCA____UCACUUUUGAUAGCCAACUAAUCGCUGCUUCUCCGCUG_ACCGGCAC .................................(((((....................))))).......................................(((.....))).. ( -6.58 = -5.35 + -1.23)
| Location | 10,248,056 – 10,248,160 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 59.67 |
| Shannon entropy | 0.78187 |
| G+C content | 0.48470 |
| Mean single sequence MFE | -40.19 |
| Consensus MFE | -7.70 |
| Energy contribution | -6.73 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.95 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.19 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.993458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
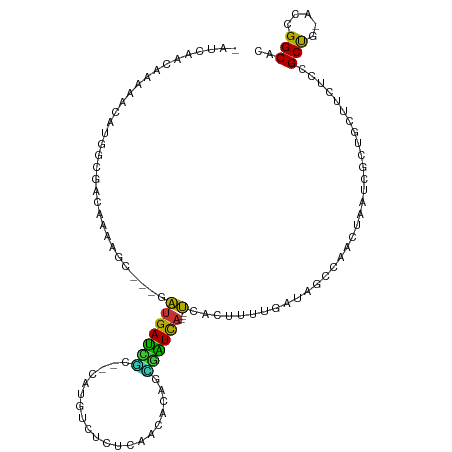
>dm3.chr2R 10248056 104 - 21146708 GUGCCGGU-CAGCGGAGAAAAAGCGAUUAUUUGGCUAUCAAAAGAUA----UGAUCACUGUGUUGAGAGACAUG--GCAAUCAUC---GCUAUUGUAACCAUAUAUUUGCUGAU- ...(((..-...)))......(((((.(((.(((.((.(((.((..(----((((..(((((((....))))))--)..))))).---.)).))))).))).))).)))))...- ( -29.00, z-score = -1.76, R) >droPer1.super_2 4614440 99 - 9036312 GUGCCUGU-CAGCGAAGAUGUAACGUAUACGUGGCG--CAAA--UGG----UGACCAAUGUAUUCACCACACUGCAGUGGUCACC---GUUUUAGCCGU--UGUUUGCGUUGC-- .(((....-..))).....((((((((.(((((((.--.(((--(((----((((((.((((..........)))).))))))))---))))..)))).--))).))))))))-- ( -36.40, z-score = -2.16, R) >dp4.chr3 4426673 99 - 19779522 GUGCCUGU-CAGCGAAGAUGUAACGUAUACGUGGCG--CAAA--UGG----UGACCAAUGUAUUCACCACACUGCAGUGGUCACC---GUUUUAGCCGU--UGUUUGCGUUGC-- .(((....-..))).....((((((((.(((((((.--.(((--(((----((((((.((((..........)))).))))))))---))))..)))).--))).))))))))-- ( -36.40, z-score = -2.16, R) >droAna3.scaffold_13266 14593597 110 - 19884421 GUGCCGUU-CAGCGGAAGGUCAACGAUUAAAUGGCAACCGAAUUUGA----UGAUUUUUGCAUUCUAACAAAUGCCGAGAUCAUCCUGUUCCUUGCCGUAAUGCUUUCGUUGAAU .....(((-((((((((((((...)))...(((((((..((((..((----(((((((.(((((......))))).)))))))))..)))).)))))))....)))))))))))) ( -39.60, z-score = -4.80, R) >droEre2.scaffold_4845 6994258 102 + 22589142 GUGCCGGU-CAGCGGAGGAGCAGCGAUUAGUUGGCUGUCAAGAGAGA----UGAUUGCUGUGUUGAAAGACAUG--GCAAUCAUA---GCUUUUGUAUCCAU-UAUUUG-UGAU- ...(((..-...))).((((((((.(.....).)))))((((((..(----(((((((((((((....))))))--)))))))).---.))))))..)))..-......-....- ( -40.00, z-score = -4.48, R) >droYak2.chr2R 10183932 103 - 21139217 GUGCCGGU-CGGCGGAGGAGCAGCGAUUAGUUGGCUACCAAGAGAGA----UGAUCGCUGUGUUGAGAAACAUG--GCAAUCAUC---GCUUUUGUACCAAU-UUUUUGCUCAU- .((((...-.)))).....(.(((((..((((((.(((.(((((.((----((((.((((((((....))))))--)).))))))---.)))))))))))))-)..))))))..- ( -42.00, z-score = -4.09, R) >droSec1.super_1 7754230 104 - 14215200 GUGCCGGU-CAGCGGAGAAAAAGCGAUUAGCUGGCUAACAAAAGAGA----UGAUCGCUCUGUUGAGAGACAUG--GCAAUCAUC---GCUUUUGUAACGAUUUAUUUGCUGAU- ......((-(((((((.(((.(((.....))).....(((((((.((----((((.(((.((((....)))).)--)).))))))---.))))))).....))).)))))))))- ( -38.00, z-score = -3.79, R) >droVir3.scaffold_12875 13551853 111 - 20611582 GUGCCGGCGCAGCGCUGCUGCAGUGCGCACAAGGCGAUCAGGCGUAGGCGUAAAGCUCUGCAGGCUUCGGCCAGCAGGGCUCGCGU--CCGCUUGUCGCCAAGUUCGAAUUGC-- ((((((..(((((...)))))..)).))))..(((((.((((((..(((((..((((((((.(((....))).)))))))).))))--)))))))))))).............-- ( -60.10, z-score = -3.06, R) >consensus GUGCCGGU_CAGCGGAGAAGCAGCGAUUACUUGGCUAUCAAAAGAGA____UGAUCACUGUAUUCAGAGACAUG__GCAAUCAUC___GCUUUUGUAGCCAUGUUUUUGUUGAU_ .(((.......))).......(((((............(((((........(((((((..................))))))).......)))))...........))))).... ( -7.70 = -6.73 + -0.97)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:22:25 2011