| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,246,178 – 10,246,281 |
| Length | 103 |
| Max. P | 0.994838 |

| Location | 10,246,178 – 10,246,281 |
|---|---|
| Length | 103 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 50.59 |
| Shannon entropy | 1.03103 |
| G+C content | 0.48490 |
| Mean single sequence MFE | -40.93 |
| Consensus MFE | 0.00 |
| Energy contribution | 0.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 0.00 |
| Mean z-score | -3.76 |
| Structure conservation index | -0.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.994838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
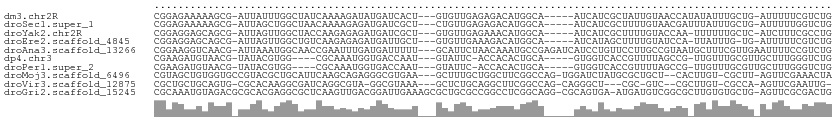
>dm3.chr2R 10246178 103 - 21146708 CGGAGAAAAAGCG-AUUAUUUGGCUAUCAAAAGAUAUGAUCACU---GUGUUGAGAGACAUGGCA-----AUCAUCGCUAUUGUAACCAUAUAUUUGCUG-AUUUUUCGUCUG (((((((..((((-(.(((.(((.((.(((.((..(((((..((---(((((....)))))))..-----)))))..)).))))).))).))).))))).-.))))))).... ( -32.60, z-score = -3.97, R) >droSec1.super_1 7754220 103 - 14215200 CGGAGAAAAAGCG-AUUAGCUGGCUAACAAAAGAGAUGAUCGCU---CUGUUGAGAGACAUGGCA-----AUCAUCGCUUUUGUAACGAUUUAUUUGCUG-AUUUUUGGUCUG (.(((((..((((-(.(((.(.(...(((((((.((((((.(((---.((((....)))).))).-----)))))).)))))))..).).))).))))).-.))))).).... ( -33.50, z-score = -3.37, R) >droYak2.chr2R 10183922 102 - 21139217 CGGAGGAGCAGCG-AUUAGUUGGCUACCAAGAGAGAUGAUCGCU---GUGUUGAGAAACAUGGCA-----AUCAUCGCUUUUGUACCAA-UUUUUUGCUC-AUCUUUCGCCUG (((((((..((((-(..((((((.(((.(((((.((((((.(((---(((((....)))))))).-----)))))).))))))))))))-))..))))).-.))))))).... ( -44.70, z-score = -5.92, R) >droEre2.scaffold_4845 6994248 101 + 22589142 CGGAGGAGCAGCG-AUUAGUUGGCUGUCAAGAGAGAUGAUUGCU---GUGUUGAAAGACAUGGCA-----AUCAUAGCUUUUGUAUCCA-UUAUUUG-UG-AUUUUUCGUCUG (((((((...(((-(.((((.((.((.((((((..(((((((((---(((((....)))))))))-----)))))..)))))))).)))-))).)))-).-.))))))).... ( -40.60, z-score = -5.52, R) >droAna3.scaffold_13266 14593587 109 - 19884421 CGGAAGGUCAACG-AUUAAAUGGCAACCGAAUUUGAUGAUUUUU---GCAUUCUAACAAAUGCCGAGAUCAUCCUGUUCCUUGCCGUAAUGCUUUCGUUGAAUUUUCCGUCUG (((((((((((((-(....(((((((..((((..(((((((((.---(((((......))))).)))))))))..)))).))))))).......)))))).)))))))).... ( -42.10, z-score = -6.67, R) >dp4.chr3 4426663 98 - 19779522 CGAAGAUGUAACG-UAUACGUGG----CGCAAAUGGUGACCAAU---GUAUUC-ACCACACUGCA-----GUGGUCACCGUUUUAGCCG-UUGUUUGCGUUGCUUUGGGUCUG .((.((.((((((-((.((((((----(..((((((((((((.(---(((...-.......))))-----.))))))))))))..))))-.))).)))))))).))...)).. ( -37.70, z-score = -3.12, R) >droPer1.super_2 4614430 98 - 9036312 CGAAGAUGUAACG-UAUACGUGG----CGCAAAUGGUGACCAAU---GUAUUC-ACCACACUGCA-----GUGGUCACCGUUUUAGCCG-UUGUUUGCGUUGCUUUGGGUCUG .((.((.((((((-((.((((((----(..((((((((((((.(---(((...-.......))))-----.))))))))))))..))))-.))).)))))))).))...)).. ( -37.70, z-score = -3.12, R) >droMoj3.scaffold_6496 7005060 105 - 26866924 CGUAGCUGUGGUGCCGUACGCUGCAUUCAAGCAGAGGGCGUGAA---GCUUUGCUGGCUUCGGCCAG-UGGAUCUAUGCGCUGCU--CACUUGU-CGCUU-AGUUCGAAACUA ((.(((((.((((.((...(((.......))).(((((((((.(---(.((..(((((....)))))-..)).)).)))))).))--)...)).-)))))-))))))...... ( -39.50, z-score = -0.69, R) >droVir3.scaffold_12875 13551854 98 - 20611582 CGCUGCUGCAGUG-CGCACAAGGCGAUCAGGCGUA-GGCGUAAA---GCUCUGCAGGCUUCGGCCAG-CAGGGCU---CGC-GUC--CGCUUGU-CGCCA-AGUUCGAAUUG- (((((...)))))-((.((..(((((.((((((..-(((((..(---(((((((.(((....))).)-)))))))---.))-)))--)))))))-)))).-.)).)).....- ( -50.60, z-score = -3.54, R) >droGri2.scaffold_15245 648703 110 + 18325388 CGCAAAUGUAGACGCGCACGAGGCGCUCAAGUUGACGGAUUGAAAGCGCUGCGCCGGCCUCGGCAGG-CGCAGUGA-AUGAUGUCGGCGCUUGUGUGCUG-AGUUCGCGACUG ........(((.((((.((..(((((.(((((.(.(.(((......(((((((((.((....)).))-))))))).-.....))).))))))).))))).-.)).)))).))) ( -50.30, z-score = -1.70, R) >consensus CGGAGAUGCAGCG_AUUACGUGGCUAUCAAAAGAGAUGAUCGAU___GUGUUGAAAGACACGGCA_____AUCAUCGCCGUUGUAGCCA_UUGUUUGCUG_AUUUUGCGUCUG ................................................................................................................. ( 0.00 = 0.00 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:22:24 2011