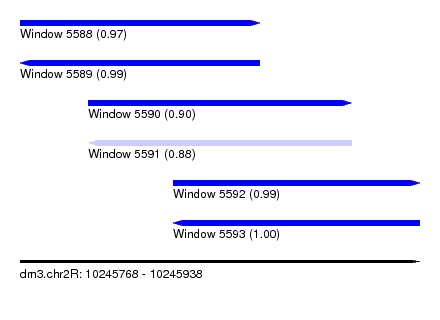
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,245,768 – 10,245,938 |
| Length | 170 |
| Max. P | 0.998744 |

| Location | 10,245,768 – 10,245,870 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 65.76 |
| Shannon entropy | 0.69271 |
| G+C content | 0.56516 |
| Mean single sequence MFE | -34.44 |
| Consensus MFE | -16.03 |
| Energy contribution | -14.70 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.974627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
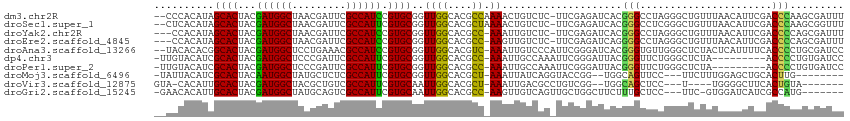
>dm3.chr2R 10245768 102 + 21146708 -UAUAUACUGGCGUGCCG-AA-GCCUGUCGCGAAAUCGCUUGGGUCGAAUGUUAAACAGCCCUAGGCCCGUGAUCUCGAA-GAGACAGUUUUGGCGUGCCAACCGC -.......((((((((((-((-((.((((.(((.(((((..(((((....(((....)))....)))))))))).)))..-..))))))))))))))))))..... ( -42.90, z-score = -4.04, R) >droSec1.super_1 7753810 102 + 14215200 -UGGAUACUGGCGUGCCG-AA-GCCUGUCGCGAAACCGCUUGGGUCGAAUGUUAAACAGCCCGAGGCCCGUGAUCUCGAA-GAGACAGUUUUAGCGUGCCAACCGC -.((....((((((((..-((-((.(((((((....)))..(((((....(((....)))....)))))...........-..))))))))..)))))))).)).. ( -34.00, z-score = -0.95, R) >droYak2.chr2R 10183517 101 + 21139217 -UAGAUUCUGGCGUGCCG-AA-ACCUGUCGCGAAAUCGCUGGGGUCGAAUGUUAAACAGCCCUAGGCCCGUGAUCUCGAA-GAGACAAUUU-GGCGUGCCAACCGC -.......((((((((((-((-...((((.(((.(((((..(((((....(((....)))....)))))))))).)))..-..)))).)))-)))))))))..... ( -37.40, z-score = -2.38, R) >droEre2.scaffold_4845 6993830 101 - 22589142 -UGGAUACUGGCGUGCCG-AA-ACCUGUCGCGAAAUCGCUGGGGUCGAAUGUUAAACAGCCCUAGGCCCCUGAUCUCGAA-GAGACAACUU-GGCGUGCCAACCGC -.((....((((((((((-(.-...((((.(((.((((..((((((....(((....)))....)))))))))).)))..-..))))..))-))))))))).)).. ( -37.10, z-score = -2.35, R) >droAna3.scaffold_13266 14593171 103 + 19884421 -UGGUUUUUGGCGUGCCUCAA-GCUUGUCGCGGGAUCGCAGGGGUGAAAAUGAGUAGAGCCCAACACCCGUGAUCCCGAAUGGGACAAUUU-GACGUGCCAACCGC -.(((...(((((((..((((-(.(((((.(((((((((..(((((....((.(.....).)).)))))))))))))).....))))))))-)))))))))))).. ( -39.60, z-score = -1.97, R) >dp4.chr3 4426264 95 + 19779522 -AUGGGAAUGACGUGCCUAAAUGCUUGCCGCGGGAUCACAGGGGUU---------AGAGCCCAGAACCCGUAAUCCCGAAUUUGGCAAUUU-GGCGUGCCAACCGC -..((...((.((((((.((.((((.....((((((.((..(((((---------.........))))))).)))))).....)))).)).-)))))).)).)).. ( -31.00, z-score = -0.31, R) >droPer1.super_2 4614031 95 + 9036312 -AUGGGAAUGACGUGCCUAAAUGCUUGCCGCGGGAUCACAGGGGUU---------AGAGCCCAGAACCCGUAAUCCCGAAUUUGGCAAUUU-GGCGUGCCAACCGC -..((...((.((((((.((.((((.....((((((.((..(((((---------.........))))))).)))))).....)))).)).-)))))).)).)).. ( -31.00, z-score = -0.31, R) >droMoj3.scaffold_6496 7004557 91 + 26866924 GUGAGAGCUGGCGUGCCU-AACGCUUACCA-GGCA-AGUGCAGCUC----------CAAAGAAGGAACUGCCA-CCGGUACCUGAUAAUUU-AGCGUGCCAACCGC (((...((((((((....-.))))).....-))).-...((((.((----------(......))).))))))-).(((((((((....))-)).)))))...... ( -29.70, z-score = -1.23, R) >droVir3.scaffold_12875 13551416 88 + 20611582 GUGAGCGCUGGCGUGCCU-AAUGCUUGCCG-AGUACAGUGAAGCCC----------CAA----GGAGCUGCCA-CCGACAGGCGUCAAUUU-AGCGUGCCAAUUGC ....(((.((((((((..-(((...((((.-.((.(.(((.(((((----------...----)).)))..))-).))).))))...))).-.))))))))..))) ( -27.30, z-score = 0.29, R) >consensus _UGGAUACUGGCGUGCCU_AA_GCUUGUCGCGGAAUCGCUGGGGUC_AA______ACAGCCCAAGACCCGUGAUCCCGAA_GAGACAAUUU_GGCGUGCCAACCGC ........((((((((...((...(((((.(((....((..(((((..................)))))))....))).....))))).))..))))))))..... (-16.03 = -14.70 + -1.33)



| Location | 10,245,768 – 10,245,870 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 65.76 |
| Shannon entropy | 0.69271 |
| G+C content | 0.56516 |
| Mean single sequence MFE | -34.23 |
| Consensus MFE | -20.51 |
| Energy contribution | -18.37 |
| Covariance contribution | -2.14 |
| Combinations/Pair | 1.74 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.994526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10245768 102 - 21146708 GCGGUUGGCACGCCAAAACUGUCUC-UUCGAGAUCACGGGCCUAGGGCUGUUUAACAUUCGACCCAAGCGAUUUCGCGACAGGC-UU-CGGCACGCCAGUAUAUA- ....(((((..(((((..(((((..-.(((((...((((.(....).))))......))))).....(((....))))))))..-))-.)))..)))))......- ( -31.30, z-score = -0.69, R) >droSec1.super_1 7753810 102 - 14215200 GCGGUUGGCACGCUAAAACUGUCUC-UUCGAGAUCACGGGCCUCGGGCUGUUUAACAUUCGACCCAAGCGGUUUCGCGACAGGC-UU-CGGCACGCCAGUAUCCA- ..(((((((.........(((((..-..(((((((.((((..(((((.((.....))))))))))..).))))))).)))))((-..-..))..)))))...)).- ( -33.30, z-score = -0.68, R) >droYak2.chr2R 10183517 101 - 21139217 GCGGUUGGCACGCC-AAAUUGUCUC-UUCGAGAUCACGGGCCUAGGGCUGUUUAACAUUCGACCCCAGCGAUUUCGCGACAGGU-UU-CGGCACGCCAGAAUCUA- ....(((((..(((-((((((((..-..(((((((.((((....(((.((.....)))))...))).).))))))).)))).))-))-.)))..)))))......- ( -28.50, z-score = 0.26, R) >droEre2.scaffold_4845 6993830 101 + 22589142 GCGGUUGGCACGCC-AAGUUGUCUC-UUCGAGAUCAGGGGCCUAGGGCUGUUUAACAUUCGACCCCAGCGAUUUCGCGACAGGU-UU-CGGCACGCCAGUAUCCA- ..(((((((..(((-.....(((((-...)))))..(..((((.(((..(((........)))))).(((....)))...))))-..-))))..)))))...)).- ( -32.60, z-score = -0.51, R) >droAna3.scaffold_13266 14593171 103 - 19884421 GCGGUUGGCACGUC-AAAUUGUCCCAUUCGGGAUCACGGGUGUUGGGCUCUACUCAUUUUCACCCCUGCGAUCCCGCGACAAGC-UUGAGGCACGCCAAAAACCA- ..(((((((.(.((-((.(((((.....(((((((.((((((.((((.....))))....)))))..).))))))).)))))..-)))).)...))))...))).- ( -39.30, z-score = -2.65, R) >dp4.chr3 4426264 95 - 19779522 GCGGUUGGCACGCC-AAAUUGCCAAAUUCGGGAUUACGGGUUCUGGGCUCU---------AACCCCUGUGAUCCCGCGGCAAGCAUUUAGGCACGUCAUUCCCAU- ..((.((((..(((-((((((((.....((((((((((((....((.....---------..)))))))))))))).))))...)))).)))..))))...))..- ( -35.20, z-score = -1.71, R) >droPer1.super_2 4614031 95 - 9036312 GCGGUUGGCACGCC-AAAUUGCCAAAUUCGGGAUUACGGGUUCUGGGCUCU---------AACCCCUGUGAUCCCGCGGCAAGCAUUUAGGCACGUCAUUCCCAU- ..((.((((..(((-((((((((.....((((((((((((....((.....---------..)))))))))))))).))))...)))).)))..))))...))..- ( -35.20, z-score = -1.71, R) >droMoj3.scaffold_6496 7004557 91 - 26866924 GCGGUUGGCACGCU-AAAUUAUCAGGUACCGG-UGGCAGUUCCUUCUUUG----------GAGCUGCACU-UGCC-UGGUAAGCGUU-AGGCACGCCAGCUCUCAC ..(((((((..(((-((.((((((((((..((-((.(((((((......)----------))))))))))-))))-))))))...))-).))..)))))))..... ( -41.80, z-score = -4.15, R) >droVir3.scaffold_12875 13551416 88 - 20611582 GCAAUUGGCACGCU-AAAUUGACGCCUGUCGG-UGGCAGCUCC----UUG----------GGGCUUCACUGUACU-CGGCAAGCAUU-AGGCACGCCAGCGCUCAC ((..(((((..(((-....((..(((.(((((-(((.(((((.----...----------))))))))))).)).-.)))...))..-.)))..))))).)).... ( -30.90, z-score = -0.75, R) >consensus GCGGUUGGCACGCC_AAAUUGUCUC_UUCGGGAUCACGGGUCUUGGGCUGU______UU_GACCCCAGCGAUUCCGCGACAAGC_UU_AGGCACGCCAGUACCCA_ ....(((((..(((....(((((.....(((((((..(((......................)))....))))))).))))).......)))..)))))....... (-20.51 = -18.37 + -2.14)


| Location | 10,245,797 – 10,245,909 |
|---|---|
| Length | 112 |
| Sequences | 10 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 66.11 |
| Shannon entropy | 0.72177 |
| G+C content | 0.53690 |
| Mean single sequence MFE | -33.73 |
| Consensus MFE | -16.61 |
| Energy contribution | -15.77 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.41 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.901038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10245797 112 + 21146708 AAAUCGCUUGGGUCGAAUGUUAAACAGCCCUAGGCCCGUGAUCUCGAA-GAGACAGUUUUGGCGUGCCAACCGCACGGAUGGCGAAUCGUUAGCCAUCGUAGUGCUAUGUGGG-- ....((((.(((((....(((....)))....))))).((.((((...-)))))).....))))..(((...((((.((((((.........))))))...))))....))).-- ( -33.70, z-score = 0.14, R) >droSec1.super_1 7753839 112 + 14215200 AAACCGCUUGGGUCGAAUGUUAAACAGCCCGAGGCCCGUGAUCUCGAA-GAGACAGUUUUAGCGUGCCAACCGCACGAAUGGCGAAUCGUUAGCCAUCGUAGUGCUAUGUGAG-- ....(((((((((....((.....))))))))(((.(((..((((...-))))........))).)))....((((..(((((.........)))))....))))...)))..-- ( -28.20, z-score = 0.65, R) >droYak2.chr2R 10183546 110 + 21139217 AAAUCGCUGGGGUCGAAUGUUAAACAGCCCUAGGCCCGUGAUCUCGAA-GAGACAAUUU-GGCGUGCCAACCGCACGGAUGGCGAAUCGUUAGCCAUCGUAGUGCUAUGUGG--- ......(((((((....((.....)))))))))(((..((.((((...-))))))....-)))...(((...((((.((((((.........))))))...))))....)))--- ( -34.60, z-score = -0.39, R) >droEre2.scaffold_4845 6993859 110 - 22589142 AAAUCGCUGGGGUCGAAUGUUAAACAGCCCUAGGCCCCUGAUCUCGAA-GAGACAACUU-GGCGUGCCAACCGCACGGAUGGCGAAUCGUUAGCCAUCGUAGUGCUAUGUGG--- ....((((((((((....(((....)))....))))))((.((((...-))))))....-))))..(((...((((.((((((.........))))))...))))....)))--- ( -35.70, z-score = -0.79, R) >droAna3.scaffold_13266 14593201 112 + 19884421 GGAUCGCAGGGGUGAAAAUGAGUAGAGCCCAACACCCGUGAUCCCGAAUGGGACAAUUU-GACGUGCCAACCGCACGGAUGGCGUUUCAGGAGCCAUCGUAGUGCCGUGUGUA-- (((((((..(((((....((.(.....).)).))))))))))))((.((((.((.....-..(((((.....)))))((((((.........))))))...)).)))).))..-- ( -40.60, z-score = -1.53, R) >dp4.chr3 4426295 104 + 19779522 GGAUCACAGGGGU---------UAGAGCCCAGAACCCGUAAUCCCGAAUUUGGCAAUUU-GGCGUGCCAACCGCACGAAUGGCGAAUCGGGAGCCAUCGUAGUGCGAUGUACAA- ((.((....((((---------....)))).)).)).(((.((((((.((((.((....-..(((((.....)))))..)).))))))))))..(((((.....))))))))..- ( -35.20, z-score = -1.22, R) >droPer1.super_2 4614062 104 + 9036312 GGAUCACAGGGGU---------UAGAGCCCAGAACCCGUAAUCCCGAAUUUGGCAAUUU-GGCGUGCCAACCGCACGAAUGGCGAAUCGGGAGCCAUCGUAGUGCGAUGUACAA- ((.((....((((---------....)))).)).)).(((.((((((.((((.((....-..(((((.....)))))..)).))))))))))..(((((.....))))))))..- ( -35.20, z-score = -1.22, R) >droMoj3.scaffold_6496 7004588 100 + 26866924 --------CAAGUGCAGCUCCAAAGAA---GGAACUGCCA--CCGGUACCUGAUAAUUU-AGCGUGCCAACCGCACGAAUGGCGAGAGCAUAGCCAUUGUAGUGCGAUGUAAUA- --------...((((((.(((......---))).))).))--).(((((((((....))-)).)))))...(((((.((((((.........))))))...)))))........- ( -36.00, z-score = -3.93, R) >droVir3.scaffold_12875 13551447 97 + 20611582 -------UACAGUGAAGCCCCA----A---GGAGCUGCCA--CCGACAGGCGUCAAUUU-AGCGUGCCAAUUGCACGAAUGGCGACAGCGUAGCCAUCGUAGUGCAAUGUG-UAC -------....(((.(((((..----.---)).)))..))--).(((....))).....-...((((..(((((((..(((((.........)))))....)))))))..)-))) ( -27.90, z-score = -0.20, R) >droGri2.scaffold_15245 648253 102 - 18325388 -------CAUGGCGAUGAUCCAC-GAA---GGAGCAAAGAAGCCAGCAACUGACAACUU-GGCGUGCCAAUUGCACGAAUGGCGACUGCAUAGCCAUCGUAGUGCAAUGUGUUC- -------..(((((.((.(((..-...---))).)).....(((((...........))-))).)))))(((((((..(((((.........)))))....)))))))......- ( -30.20, z-score = -0.36, R) >consensus AAAUCGCUGGGGUCGAAUGUUAAAGAGCCCGAAGCCCGUAAUCCCGAA_GAGACAAUUU_GGCGUGCCAACCGCACGAAUGGCGAAUCGGUAGCCAUCGUAGUGCUAUGUGGA__ .........((((....................))))...............(((.......(((((.....))))).(((((.........)))))..........)))..... (-16.61 = -15.77 + -0.84)
| Location | 10,245,797 – 10,245,909 |
|---|---|
| Length | 112 |
| Sequences | 10 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 66.11 |
| Shannon entropy | 0.72177 |
| G+C content | 0.53690 |
| Mean single sequence MFE | -32.37 |
| Consensus MFE | -15.59 |
| Energy contribution | -14.79 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.40 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.883384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10245797 112 - 21146708 --CCCACAUAGCACUACGAUGGCUAACGAUUCGCCAUCCGUGCGGUUGGCACGCCAAAACUGUCUC-UUCGAGAUCACGGGCCUAGGGCUGUUUAACAUUCGACCCAAGCGAUUU --....((..((((...((((((.........)))))).))))...))((..(((......(((((-...)))))....)))...(((.((.........)).)))..))..... ( -28.60, z-score = 0.20, R) >droSec1.super_1 7753839 112 - 14215200 --CUCACAUAGCACUACGAUGGCUAACGAUUCGCCAUUCGUGCGGUUGGCACGCUAAAACUGUCUC-UUCGAGAUCACGGGCCUCGGGCUGUUUAACAUUCGACCCAAGCGGUUU --......((((.....((((((.........)))))).((((.....))))))))((((((((((-...))).....(((..(((((.((.....))))))))))..))))))) ( -29.80, z-score = -0.11, R) >droYak2.chr2R 10183546 110 - 21139217 ---CCACAUAGCACUACGAUGGCUAACGAUUCGCCAUCCGUGCGGUUGGCACGCC-AAAUUGUCUC-UUCGAGAUCACGGGCCUAGGGCUGUUUAACAUUCGACCCCAGCGAUUU ---...((..((((...((((((.........)))))).))))...))((..(((-.....(((((-...)))))....)))...(((..(((........)))))).))..... ( -29.00, z-score = 0.10, R) >droEre2.scaffold_4845 6993859 110 + 22589142 ---CCACAUAGCACUACGAUGGCUAACGAUUCGCCAUCCGUGCGGUUGGCACGCC-AAGUUGUCUC-UUCGAGAUCAGGGGCCUAGGGCUGUUUAACAUUCGACCCCAGCGAUUU ---.......((((...((((((.........)))))).))))((((((((.(((-.....(((((-(........))))))....))))))........))))).......... ( -30.30, z-score = 0.30, R) >droAna3.scaffold_13266 14593201 112 - 19884421 --UACACACGGCACUACGAUGGCUCCUGAAACGCCAUCCGUGCGGUUGGCACGUC-AAAUUGUCCCAUUCGGGAUCACGGGUGUUGGGCUCUACUCAUUUUCACCCCUGCGAUCC --..((.((.((((...((((((.........)))))).)))).))))(((....-.....(((((....)))))...(((((.((((.....))))....))))).)))..... ( -36.30, z-score = -1.14, R) >dp4.chr3 4426295 104 - 19779522 -UUGUACAUCGCACUACGAUGGCUCCCGAUUCGCCAUUCGUGCGGUUGGCACGCC-AAAUUGCCAAAUUCGGGAUUACGGGUUCUGGGCUCUA---------ACCCCUGUGAUCC -......(((((((...((((((.........)))))).)))))))(((((....-....)))))......((((((((((....((......---------.)))))))))))) ( -36.00, z-score = -1.77, R) >droPer1.super_2 4614062 104 - 9036312 -UUGUACAUCGCACUACGAUGGCUCCCGAUUCGCCAUUCGUGCGGUUGGCACGCC-AAAUUGCCAAAUUCGGGAUUACGGGUUCUGGGCUCUA---------ACCCCUGUGAUCC -......(((((((...((((((.........)))))).)))))))(((((....-....)))))......((((((((((....((......---------.)))))))))))) ( -36.00, z-score = -1.77, R) >droMoj3.scaffold_6496 7004588 100 - 26866924 -UAUUACAUCGCACUACAAUGGCUAUGCUCUCGCCAUUCGUGCGGUUGGCACGCU-AAAUUAUCAGGUACCGG--UGGCAGUUCC---UUCUUUGGAGCUGCACUUG-------- -.....(((((.(((..((((((.........))))))(((((.....)))))..-.........)))..)))--))((((((((---......)))))))).....-------- ( -33.40, z-score = -2.65, R) >droVir3.scaffold_12875 13551447 97 - 20611582 GUA-CACAUUGCACUACGAUGGCUACGCUGUCGCCAUUCGUGCAAUUGGCACGCU-AAAUUGACGCCUGUCGG--UGGCAGCUCC---U----UGGGGCUUCACUGUA------- ...-...(((((((...((((((.((...)).)))))).))))))).((((.((.-........)).))))((--(((.(((((.---.----..))))))))))...------- ( -32.30, z-score = -0.90, R) >droGri2.scaffold_15245 648253 102 + 18325388 -GAACACAUUGCACUACGAUGGCUAUGCAGUCGCCAUUCGUGCAAUUGGCACGCC-AAGUUGUCAGUUGCUGGCUUCUUUGCUCC---UUC-GUGGAUCAUCGCCAUG------- -......(((((((...((((((.........)))))).)))))))((((..((.-(((..(((((...)))))..))).))(((---...-..))).....))))..------- ( -32.00, z-score = -1.10, R) >consensus __ACCACAUAGCACUACGAUGGCUAACGAUUCGCCAUUCGUGCGGUUGGCACGCC_AAAUUGUCAC_UUCGGGAUCACGGGCUCCGGGCUCUUUAACAUUCCACCCCAGCGAUUU ..........((.....((((((.........)))))).((((.....))))))........................(((......................)))......... (-15.59 = -14.79 + -0.80)

| Location | 10,245,833 – 10,245,938 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 66.74 |
| Shannon entropy | 0.72660 |
| G+C content | 0.45664 |
| Mean single sequence MFE | -30.46 |
| Consensus MFE | -13.74 |
| Energy contribution | -14.01 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.986832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
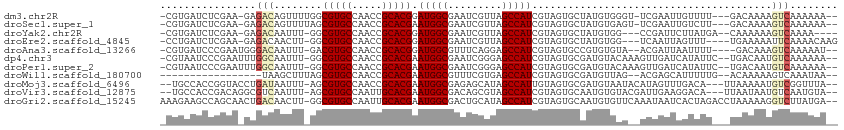
>dm3.chr2R 10245833 105 + 21146708 -CGUGAUCUCGAA-GAGACAGUUUUGGCGUGCCAACCGCACGGAUGGCGAAUCGUUAGCCAUCGUAGUGCUAUGUGGGU-UCGAAUUGUUUU---GACAAAAGUCAAAAAA-- -.....((((...-))))((((((..((.(((.....((((.((((((.........))))))...))))...))).))-..))))))((((---(((....)))))))..-- ( -30.30, z-score = -0.97, R) >droSec1.super_1 7753875 105 + 14215200 -CGUGAUCUCGAA-GAGACAGUUUUAGCGUGCCAACCGCACGAAUGGCGAAUCGUUAGCCAUCGUAGUGCUAUGUGAGU-UCGAAUUGUCUU---GACAAAAGUCAAAAAA-- -..((((......-((((((((((.(((..((....(((((((.((((.........)))))))).)))....))..))-).))))))))))---.......)))).....-- ( -28.52, z-score = -1.31, R) >droYak2.chr2R 10183582 101 + 21139217 -CGUGAUCUCGAA-GAGACAAUUU-GGCGUGCCAACCGCACGGAUGGCGAAUCGUUAGCCAUCGUAGUGCUAUGUGG---CCGAUUCUUAUGA--CAAAAAAGUCAAAA---- -..((.((((...-)))))).(((-(((..((((.((....)).))))((((((...((((.((((....)))))))---)))))))......--.......)))))).---- ( -31.50, z-score = -1.95, R) >droEre2.scaffold_4845 6993895 103 - 22589142 -CCUGAUCUCGAA-GAGACAACUU-GGCGUGCCAACCGCACGGAUGGCGAAUCGUUAGCCAUCGUAGUGCUAUGUGG---UCAAUUAGUUU----UGAAAAAUUCAAAACAAG -..(((((.((.(-(..((.((..-..(((((.....)))))((((((.........)))))))).)).)).)).))---)))....((((----(((.....)))))))... ( -31.20, z-score = -2.49, R) >droAna3.scaffold_13266 14593237 103 + 19884421 -CGUGAUCCCGAAUGGGACAAUUU-GACGUGCCAACCGCACGGAUGGCGUUUCAGGAGCCAUCGUAGUGCCGUGUGUA--ACGAUUAAUUUU----GACAAAGUCAAAAAU-- -..(((((.((.((((.((.....-..(((((.....)))))((((((.........))))))...)).)))).))..--..))))).((((----(((...)))))))..-- ( -30.60, z-score = -1.29, R) >dp4.chr3 4426322 107 + 19779522 -CGUAAUCCCGAAUUUGGCAAUUU-GGCGUGCCAACCGCACGAAUGGCGAAUCGGGAGCCAUCGUAGUGCGAUGUACAAAGUUGAUCAUAUUC--UGACAAUGUCAAAAAA-- -.(((.((((((.((((.((....-..(((((.....)))))..)).))))))))))..(((((.....))))))))...((((.(((.....--))))))).........-- ( -30.40, z-score = -1.11, R) >droPer1.super_2 4614089 107 + 9036312 -CGUAAUCCCGAAUUUGGCAAUUU-GGCGUGCCAACCGCACGAAUGGCGAAUCGGGAGCCAUCGUAGUGCGAUGUACAAAGUUGAUCAUAUUC--UGACAAUGUCAAAAAA-- -.(((.((((((.((((.((....-..(((((.....)))))..)).))))))))))..(((((.....))))))))...((((.(((.....--))))))).........-- ( -30.40, z-score = -1.11, R) >droWil1.scaffold_180700 2979683 90 - 6630534 -----------------UAAGCUUUAGCGUGCCAACCGCACGAAUGGCGUUUCGUGAGCCAUCGUAGUGCGAUGUUAG--ACGAGCAUUUUUG--ACAAAAAGUCAAAUAA-- -----------------...(((....(((((.....)))))...)))(((.(((.(((.((((.....)))))))..--))))))...((((--((.....))))))...-- ( -25.80, z-score = -1.68, R) >droMoj3.scaffold_6496 7004612 105 + 26866924 --UGCCACCGGUACCUGAUAAUUU-AGCGUGCCAACCGCACGAAUGGCGAGAGCAUAGCCAUUGUAGUGCGAUGUAAUACAUAGUUUGACA---UUAAAAAUGUCGGUUUA-- --.(((...(((((((((....))-)).)))))...(((((.((((((.........))))))...)))))................((((---(.....))))))))...-- ( -33.00, z-score = -2.77, R) >droVir3.scaffold_12875 13551468 105 + 20611582 --UGCCACCGACAGGCGUCAAUUU-AGCGUGCCAAUUGCACGAAUGGCGACAGCGUAGCCAUCGUAGUGCAAUGUGUACGAUUGAAGGACA---UUAAUAAUGUCAAUGUA-- --.(((.......))).....(((-(((((((..(((((((..(((((.........)))))....)))))))..))))).))))).((((---(.....)))))......-- ( -32.30, z-score = -1.26, R) >droGri2.scaffold_15245 648277 110 - 18325388 AAAGAAGCCAGCAACUGACAACUU-GGCGUGCCAAUUGCACGAAUGGCGACUGCAUAGCCAUCGUAGUGCAAUGUGUUCAAAUAAUCACUAGACCUAAAAAGGUCUUAUGA-- ...((((((((...........))-)))......(((((((..(((((.........)))))....)))))))...)))......(((..((((((....))))))..)))-- ( -31.00, z-score = -1.85, R) >consensus _CGUGAUCCCGAA_GAGACAAUUU_GGCGUGCCAACCGCACGAAUGGCGAAUCGUUAGCCAUCGUAGUGCGAUGUGAA__ACGAAUCGUUUU___UAAAAAAGUCAAAAAA__ ................(((........(((((.....))))).(((((.........)))))........................................)))........ (-13.74 = -14.01 + 0.27)


| Location | 10,245,833 – 10,245,938 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 66.74 |
| Shannon entropy | 0.72660 |
| G+C content | 0.45664 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -14.73 |
| Energy contribution | -14.34 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.47 |
| SVM RNA-class probability | 0.998744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10245833 105 - 21146708 --UUUUUUGACUUUUGUC---AAAACAAUUCGA-ACCCACAUAGCACUACGAUGGCUAACGAUUCGCCAUCCGUGCGGUUGGCACGCCAAAACUGUCUC-UUCGAGAUCACG- --..(((((((....)))---))))...(((((-(...(((..((((...((((((.........)))))).))))..((((....))))...)))...-))))))......- ( -29.60, z-score = -3.21, R) >droSec1.super_1 7753875 105 - 14215200 --UUUUUUGACUUUUGUC---AAGACAAUUCGA-ACUCACAUAGCACUACGAUGGCUAACGAUUCGCCAUUCGUGCGGUUGGCACGCUAAAACUGUCUC-UUCGAGAUCACG- --..(((((((....)))---))))...(((((-(...(((((((.....((((((.........)))))).((((.....))))))))....)))...-))))))......- ( -27.70, z-score = -2.22, R) >droYak2.chr2R 10183582 101 - 21139217 ----UUUUGACUUUUUUG--UCAUAAGAAUCGG---CCACAUAGCACUACGAUGGCUAACGAUUCGCCAUCCGUGCGGUUGGCACGCC-AAAUUGUCUC-UUCGAGAUCACG- ----...((((......)--))).......(((---((((...((((...((((((.........)))))).)))).).)))).))..-.....(((((-...)))))....- ( -27.40, z-score = -1.67, R) >droEre2.scaffold_4845 6993895 103 + 22589142 CUUGUUUUGAAUUUUUCA----AAACUAAUUGA---CCACAUAGCACUACGAUGGCUAACGAUUCGCCAUCCGUGCGGUUGGCACGCC-AAGUUGUCUC-UUCGAGAUCAGG- ((((((((((.....)))----)))).....((---(.((...((((...((((((.........)))))).))))..((((....))-)))).)))..-...)))......- ( -27.70, z-score = -1.84, R) >droAna3.scaffold_13266 14593237 103 - 19884421 --AUUUUUGACUUUGUC----AAAAUUAAUCGU--UACACACGGCACUACGAUGGCUCCUGAAACGCCAUCCGUGCGGUUGGCACGUC-AAAUUGUCCCAUUCGGGAUCACG- --...((((((..((((----((.......(((--.....)))((((...((((((.........)))))).))))..)))))).)))-)))..(((((....)))))....- ( -31.40, z-score = -2.55, R) >dp4.chr3 4426322 107 - 19779522 --UUUUUUGACAUUGUCA--GAAUAUGAUCAACUUUGUACAUCGCACUACGAUGGCUCCCGAUUCGCCAUUCGUGCGGUUGGCACGCC-AAAUUGCCAAAUUCGGGAUUACG- --..(((((((...))))--))).............((((((((.....)))))..((((((.(((((....).))))((((((....-....))))))..)))))).))).- ( -29.80, z-score = -1.72, R) >droPer1.super_2 4614089 107 - 9036312 --UUUUUUGACAUUGUCA--GAAUAUGAUCAACUUUGUACAUCGCACUACGAUGGCUCCCGAUUCGCCAUUCGUGCGGUUGGCACGCC-AAAUUGCCAAAUUCGGGAUUACG- --..(((((((...))))--))).............((((((((.....)))))..((((((.(((((....).))))((((((....-....))))))..)))))).))).- ( -29.80, z-score = -1.72, R) >droWil1.scaffold_180700 2979683 90 + 6630534 --UUAUUUGACUUUUUGU--CAAAAAUGCUCGU--CUAACAUCGCACUACGAUGGCUCACGAAACGCCAUUCGUGCGGUUGGCACGCUAAAGCUUA----------------- --...((((((.....))--))))...((((((--(((((..(((((...((((((.........)))))).)))))))))).)))....)))...----------------- ( -27.60, z-score = -3.13, R) >droMoj3.scaffold_6496 7004612 105 - 26866924 --UAAACCGACAUUUUUAA---UGUCAAACUAUGUAUUACAUCGCACUACAAUGGCUAUGCUCUCGCCAUUCGUGCGGUUGGCACGCU-AAAUUAUCAGGUACCGGUGGCA-- --...((((((((.....)---)))).......(((((...((((((...((((((.........)))))).))))))((((....))-)).......))))).)))....-- ( -25.70, z-score = -0.83, R) >droVir3.scaffold_12875 13551468 105 - 20611582 --UACAUUGACAUUAUUAA---UGUCCUUCAAUCGUACACAUUGCACUACGAUGGCUACGCUGUCGCCAUUCGUGCAAUUGGCACGCU-AAAUUGACGCCUGUCGGUGGCA-- --......(((((.....)---)))).......((((..(((((.....)))))..))))..((((((...(((((.....)))))..-.....(((....))))))))).-- ( -28.80, z-score = -1.03, R) >droGri2.scaffold_15245 648277 110 + 18325388 --UCAUAAGACCUUUUUAGGUCUAGUGAUUAUUUGAACACAUUGCACUACGAUGGCUAUGCAGUCGCCAUUCGUGCAAUUGGCACGCC-AAGUUGUCAGUUGCUGGCUUCUUU --((((.((((((....)))))).))))...............((...((((((((.........)))).))))(((((((((((...-..).))))))))))..))...... ( -34.60, z-score = -2.52, R) >consensus __UUUUUUGACUUUUUUA___AAAACGAUUCGU__UACACAUAGCACUACGAUGGCUAACGAUUCGCCAUUCGUGCGGUUGGCACGCC_AAAUUGUCAC_UUCGGGAUCACG_ ...........................................((.....((((((.........)))))).((((.....)))))).......................... (-14.73 = -14.34 + -0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:22:23 2011