
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,211,338 – 10,211,428 |
| Length | 90 |
| Max. P | 0.505973 |

| Location | 10,211,338 – 10,211,428 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 72.96 |
| Shannon entropy | 0.51297 |
| G+C content | 0.43183 |
| Mean single sequence MFE | -22.52 |
| Consensus MFE | -13.63 |
| Energy contribution | -13.64 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.07 |
| Mean z-score | -0.77 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.505973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10211338 90 + 21146708 ----------------AUAGCGAAGGCAAAGUAA-AGGGCCCCGUGGGACAGUUGCUGAUUUAAAUUC-GUGUUAGAGAGACAAAAUAAUUACGGCAAUUGGGCAGGA---------- ----------------...((....)).......-...(((((...)).(((((((((..........-.((((.....)))).........))))))))))))....---------- ( -19.85, z-score = -0.47, R) >droPer1.super_2 4429517 111 + 9036312 AAAGGGCC----CAAAAAAAAAAAAAAAUAGC---UGUGCCCCCUGGGACAGUUGCUGAUUUAAAUUCCGUGUUAGAGAGACAAAAUAAUUGCGGCAAUUGGUCGUAGAAGCCAGAAA ...((((.----((..................---)).)))).(((.(((((((((((.....((((...((((.....))))....)))).)))))))).))).))).......... ( -24.17, z-score = -0.48, R) >dp4.chr3 4242277 110 + 19779522 AAAGGGCC----CAAAAAAAAAAAAAA-UAGC---UGUGCCCCCUGGGACAGUUGCUGAUUUAAAUUCCGUGUUAGAGAGACAAAAUAAUUGCGGCAAUUGGUCGUAGAAGCCAGAAA ...((((.----((.............-....---)).)))).(((.(((((((((((.....((((...((((.....))))....)))).)))))))).))).))).......... ( -24.23, z-score = -0.52, R) >droAna3.scaffold_13266 14559634 99 + 19884421 ----------------AAAGCAGAAAAAUAGCAA-AGGGCCCCGUGGGACAGUUGCUGAUUUAAAUUC-GUGUUAGAGAGACACAAUAAUUACGGCAAUUGGCCA-AAAAGCAGUAAA ----------------..............((..-..(((((....)).(((((((((......(((.-(((((.....)))))))).....)))))))))))).-....))...... ( -23.40, z-score = -1.63, R) >droEre2.scaffold_4845 6959809 90 - 22589142 ----------------ACAGCGAAGGCAAAGUAA-AGGGCCCCGUGGGACAGUUGCUGAUUUAAAUUC-GUGUUAGAGAGACAAAAUAAUUACGGCAAUUGGGCAGGA---------- ----------------...((....)).......-...(((((...)).(((((((((..........-.((((.....)))).........))))))))))))....---------- ( -19.85, z-score = -0.33, R) >droYak2.chr2R 10148945 90 + 21139217 ----------------ACGGCGAAGGCAAAGUAA-AGGGCCCCGUGGGACAGUUGCUGAUUUAAGUUC-GUGUUAGAGAGCCAAAAUAAUUACGGCAAUUGGGCAGGA---------- ----------------((.((....))...))..-...(((((...)).(((((((((((((..((((-........))))..)))).....))))))))))))....---------- ( -21.80, z-score = 0.17, R) >droSec1.super_1 7719935 90 + 14215200 ----------------ACAGCGAAGGCAAAGUAA-AGGGCCCCGUGGGACAGUUGCUGAUUUAAAUUC-GUGUUAGAGAGACAAAAUAAUUACGGCAAUUGGGCAGGA---------- ----------------...((....)).......-...(((((...)).(((((((((..........-.((((.....)))).........))))))))))))....---------- ( -19.85, z-score = -0.33, R) >droSim1.chr2R 8975103 90 + 19596830 ----------------ACAGCGAAGGCAAAGUAA-AGGGCCCCGUGGGACAGUUGCUGAUUUAAAUUC-GUGUUAGAGAGACAAAAUAAUUACGGCAAUUGGGCAGGA---------- ----------------...((....)).......-...(((((...)).(((((((((..........-.((((.....)))).........))))))))))))....---------- ( -19.85, z-score = -0.33, R) >droWil1.scaffold_180700 2923353 116 - 6630534 AAAGGGCCGUAGUAAACGAAAUGAAAGAUUGCCAUUGCCCCCUCUGGGACAGUUGCUGAUUUAAAUUC-GUGUUAGAGAGACAAAAUAAUUACAGCAAUUGGCCA-AAAGGAAAAAAA ...((((.((.((((.(.........).)))).)).))))(((...((.(((((((((..........-.((((.....)))).........))))))))).)).-..)))....... ( -28.75, z-score = -2.11, R) >droVir3.scaffold_12875 13507377 87 + 20611582 -------------------AAAGGGCCCCAUU------GCCCCGUGGGACAGUUGCUGAUUUAAAUUC-GUGUUAGAGAGACAAAAUAAUUACGGCAAUUGGCCAAAAACAAA----- -------------------....((((((((.------.....))))).(((((((((..........-.((((.....)))).........)))))))))))).........----- ( -23.25, z-score = -1.43, R) >droMoj3.scaffold_6496 6951704 98 + 26866924 ------------------AAAAGGGCCCCAGUCG-AAUGGCCCGUGGGACAGUUGCUGAUUUAAAUUC-GUGUUAGAGAGACAAAAUAAUUACGGCAAUUGGGCAAAAAAAAAAGAAG ------------------....(((((.......-...)))))...(..(((((((((..........-.((((.....)))).........)))))))))..).............. ( -22.75, z-score = -1.04, R) >consensus ________________ACAGCAAAGGCAAAGUAA_AGGGCCCCGUGGGACAGUUGCUGAUUUAAAUUC_GUGUUAGAGAGACAAAAUAAUUACGGCAAUUGGGCAGAAA_________ ........................................((....)).(((((((((............((((.....)))).........)))))))))................. (-13.63 = -13.64 + 0.01)

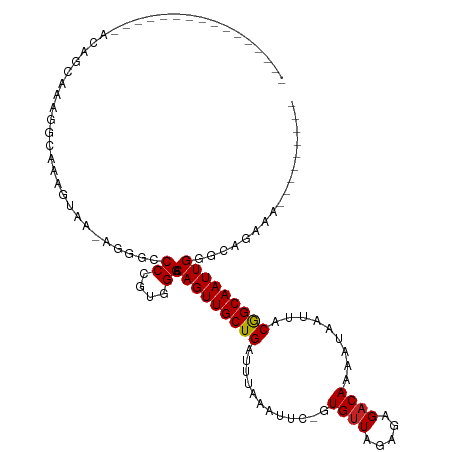
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:22:16 2011