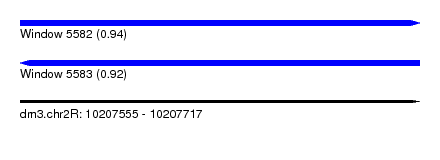
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,207,555 – 10,207,717 |
| Length | 162 |
| Max. P | 0.938747 |

| Location | 10,207,555 – 10,207,717 |
|---|---|
| Length | 162 |
| Sequences | 5 |
| Columns | 176 |
| Reading direction | forward |
| Mean pairwise identity | 76.73 |
| Shannon entropy | 0.40559 |
| G+C content | 0.45989 |
| Mean single sequence MFE | -53.88 |
| Consensus MFE | -30.46 |
| Energy contribution | -30.74 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
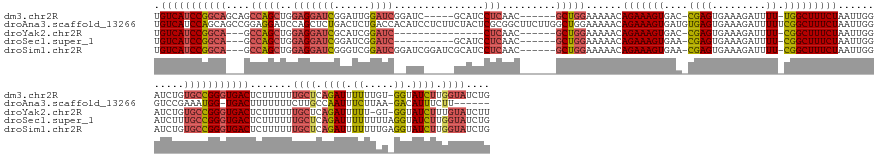
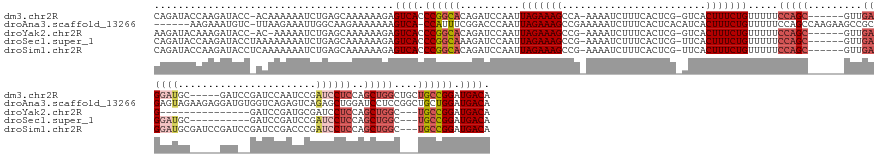
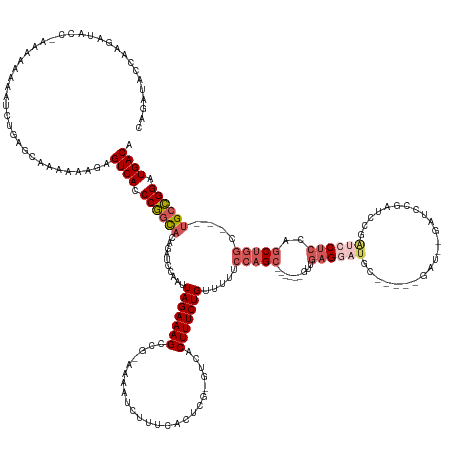
>dm3.chr2R 10207555 162 + 21146708 UGUCAUCCGGCAGCAGCCAGCUGGAGGAUCGGAUUGGAUCGGAUC-----GCAUCCUCAAC------GCUGGAAAAACAGAAAGUGAC-CGAGUGAAAGAUUUU-UGGCUUUCUAAUUGGAUCUGUGCCGGGUGACUCUUUUUUGCUCAGAUUUUUUGU-GGUAUCUUGGUAUCUG .(((((((((((.((((((((..((((((.(((((......))))-----.)))))))...------)))))...............(-(((..(((((.....-...)))))...))))..)))))))))))))).......((((.((((.......-...)))).)))).... ( -56.10, z-score = -2.13, R) >droAna3.scaffold_13266 14556558 168 + 19884421 UGUCAUCCAGCAGCCGGAGGAUCCAGCUCUGACUCUGACCACAUCCUCUUCUACUCGCGGCUUCUUGGCUGGAAAAACAGAAAGUGAUGUGAGUGAAAGAUUUUUCGGCUUUCUAAUUGGGUCCGAAAUGG-UGACUUUUUUUCUUGCCAAUUUCUUAA-GACAUUUCUU------ ((((((((((.(((((((((((((((........)))((((((((((.((((..((.(((((....))))))).....)))))).)))))).))....))))))))))))......))))))..(((((((-..(.........)..)).)))))....-))))......------ ( -47.00, z-score = -0.64, R) >droYak2.chr2R 10145199 148 + 21139217 UGUCAUCCGGCA---GCCAGCUGGAGGAUCGCAUCGGAUC---------------CUCAAC------GCUGGAAAAACAGAAAGUGAC-CGAGUGAAAGAUUUU-CGGCUUUCUAAUUGGAUCUGUGCCGGGUGACUCUUUUUUGCUCAGAUUUUU-GU-GGUAUCUUUGUAUCUU .((((((((((.---.(((((..(((((((......))))---------------)))...------)))))....(((((......(-(((..(((((.....-...)))))...)))).))))))))))))))).......(((..((((....-..-...))))..))).... ( -54.50, z-score = -3.39, R) >droSec1.super_1 7714449 155 + 14215200 UGUCAUCCGGCA---GCCAGCUGGAGGAUCGGAUCGGAUC----------GCAUCCUCAAC------GCUGGAAAAACAGAAAGUGAA-CGAGUGAAAGAUUUU-CGGCUUUCUAAUUGGAUCUUUGCCGGGUGACUCUUUUUUGCUCAGAUUUUUUUUAGGUAUCUUGGUAUCUG .(((((((((((---((((((..(((((((((......))----------).))))))...------)))))......(((((((...-((((.........))-)))))))))..........))))))))))))......................(((((((....))))))) ( -53.00, z-score = -2.49, R) >droSim1.chr2R 8970167 165 + 19596830 UGUCAUCCGGCA---GCCAGCUGGAGGAUCGGGUCGGAUCGGAUCGGAUCGCAUCCUCAAC------GCUGGAAAAACAGAAAGUGAA-CGAGUGAAAGAUUUU-CGGCUUUCUAAUUGGAUCUGUGCCGGGUGACUCUUUUUUGCUCAGAUUUUUUUGAGGUAUCUUGGUAUCUG .(((((((((((---.(((((..((((((.(((((.(((...))).)))).)))))))...------)))))......(((((((...-((((.........))-)))))))))...........))))))))))).........((((((....))))))((((....))))... ( -58.80, z-score = -2.24, R) >consensus UGUCAUCCGGCA___GCCAGCUGGAGGAUCGGAUCGGAUC__AUC_____GCAUCCUCAAC______GCUGGAAAAACAGAAAGUGAA_CGAGUGAAAGAUUUU_CGGCUUUCUAAUUGGAUCUGUGCCGGGUGACUCUUUUUUGCUCAGAUUUUUUGU_GGUAUCUUGGUAUCUG .(((((((((((....((....))..........((((((............................(((......))).........(((..(((((.........)))))...)))))))))))))))))))).......((((.((((.((.....)).)))).)))).... (-30.46 = -30.74 + 0.28)
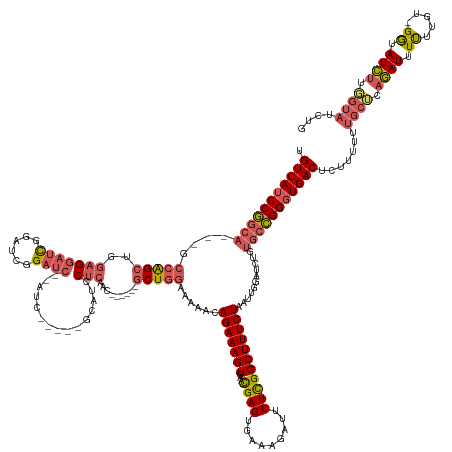
| Location | 10,207,555 – 10,207,717 |
|---|---|
| Length | 162 |
| Sequences | 5 |
| Columns | 176 |
| Reading direction | reverse |
| Mean pairwise identity | 76.73 |
| Shannon entropy | 0.40559 |
| G+C content | 0.45989 |
| Mean single sequence MFE | -44.22 |
| Consensus MFE | -24.52 |
| Energy contribution | -23.68 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.915558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10207555 162 - 21146708 CAGAUACCAAGAUACC-ACAAAAAAUCUGAGCAAAAAAGAGUCACCCGGCACAGAUCCAAUUAGAAAGCCA-AAAAUCUUUCACUCG-GUCACUUUCUGUUUUUCCAGC------GUUGAGGAUGC-----GAUCCGAUCCAAUCCGAUCCUCCAGCUGGCUGCUGCCGGAUGACA (((((...........-.......)))))...........((((.(((((((((.......(((((((((.-..............)-)...))))))).....(((((------...(((((((.-----(((........)))).))))))..)))))))).)))))).)))). ( -45.23, z-score = -2.52, R) >droAna3.scaffold_13266 14556558 168 - 19884421 ------AAGAAAUGUC-UUAAGAAAUUGGCAAGAAAAAAAGUCA-CCAUUUCGGACCCAAUUAGAAAGCCGAAAAAUCUUUCACUCACAUCACUUUCUGUUUUUCCAGCCAAGAAGCCGCGAGUAGAAGAGGAUGUGGUCAGAGUCAGAGCUGGAUCCUCCGGCUGCUGGAUGACA ------..((((((((-..........)))).............-...((((((..............))))))....))))...((((((..((((((...((((.((......)).).)))))))))..))))))((((...((((((((((.....)))))).)))).)))). ( -42.74, z-score = -0.02, R) >droYak2.chr2R 10145199 148 - 21139217 AAGAUACAAAGAUACC-AC-AAAAAUCUGAGCAAAAAAGAGUCACCCGGCACAGAUCCAAUUAGAAAGCCG-AAAAUCUUUCACUCG-GUCACUUUCUGUUUUUCCAGC------GUUGAG---------------GAUCCGAUGCGAUCCUCCAGCUGGC---UGCCGGAUGACA .........((((...-..-....))))............((((.((((((..........((((((((((-(...........)))-)...))))))).....(((((------...(((---------------((((......)))))))..))))).---)))))).)))). ( -45.60, z-score = -3.35, R) >droSec1.super_1 7714449 155 - 14215200 CAGAUACCAAGAUACCUAAAAAAAAUCUGAGCAAAAAAGAGUCACCCGGCAAAGAUCCAAUUAGAAAGCCG-AAAAUCUUUCACUCG-UUCACUUUCUGUUUUUCCAGC------GUUGAGGAUGC----------GAUCCGAUCCGAUCCUCCAGCUGGC---UGCCGGAUGACA (((((...................)))))...........((((.((((((..........(((((((.((-(...........)))-....))))))).....(((((------...((((((.(----------(........))))))))..))))).---)))))).)))). ( -43.41, z-score = -2.79, R) >droSim1.chr2R 8970167 165 - 19596830 CAGAUACCAAGAUACCUCAAAAAAAUCUGAGCAAAAAAGAGUCACCCGGCACAGAUCCAAUUAGAAAGCCG-AAAAUCUUUCACUCG-UUCACUUUCUGUUUUUCCAGC------GUUGAGGAUGCGAUCCGAUCCGAUCCGACCCGAUCCUCCAGCUGGC---UGCCGGAUGACA ...............((((........)))).........((((.((((((..........(((((((.((-(...........)))-....))))))).....(((((------...((((((.((.((.(((...))).))..))))))))..))))).---)))))).)))). ( -44.10, z-score = -2.09, R) >consensus CAGAUACCAAGAUACC_AAAAAAAAUCUGAGCAAAAAAGAGUCACCCGGCACAGAUCCAAUUAGAAAGCCG_AAAAUCUUUCACUCG_GUCACUUUCUGUUUUUCCAGC______GUUGAGGAUGC_____GAU__GAUCCGAUCCGAUCCUCCAGCUGGC___UGCCGGAUGACA ........................................((((.((((((..((((....(((((((........................)))))))...(((((((......)))).))).............))))............((....))....)))))).)))). (-24.52 = -23.68 + -0.84)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:22:15 2011