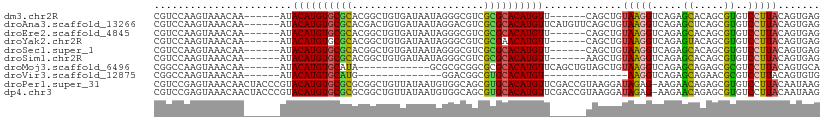
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,162,610 – 10,162,709 |
| Length | 99 |
| Max. P | 0.979101 |

| Location | 10,162,610 – 10,162,709 |
|---|---|
| Length | 99 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 77.05 |
| Shannon entropy | 0.46087 |
| G+C content | 0.50990 |
| Mean single sequence MFE | -30.87 |
| Consensus MFE | -18.23 |
| Energy contribution | -18.59 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.974534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
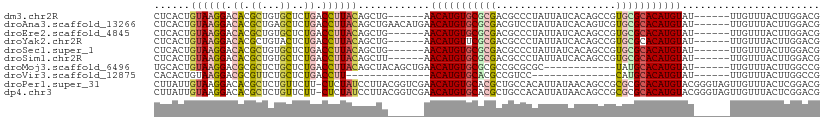
>dm3.chr2R 10162610 99 + 21146708 CUCACUGUAAGGACACGCUGUGCUCUGACCUUACAGCUG------AACAUGUGCGCGACGCCCUAUUAUCACAGCCGUGCGCACAUGUAU------UUGUUUACUUGGACG .(((((((((((.((.((...))..)).)))))))).))------)((((((((((.(((..((........)).)))))))))))))..------..((((....)))). ( -32.90, z-score = -2.73, R) >droAna3.scaffold_13266 14512666 105 + 19884421 CUCACUGUAAGGACACGCUGAGCUCUGACCUUACAGCUGAACAUGAACAUGUGCGCGACGUCCUAUUAUCACAGUCGUGCGCACAUGUAU------UUGUUUACUUGGACG ....((((((((.((.((...))..)).)))))))).((((((...((((((((((.(((.(...........).)))))))))))))..------.))))))........ ( -35.10, z-score = -2.95, R) >droEre2.scaffold_4845 6909901 99 - 22589142 CUCACUGUAAGGACACGCUGUGCUCUGACCUUACAGCUG------AACAUGUGCGCGACGCCCUAUUAUCACAGCCGUGCGCACAUGUAU------UUGUUUACUUGGACG .(((((((((((.((.((...))..)).)))))))).))------)((((((((((.(((..((........)).)))))))))))))..------..((((....)))). ( -32.90, z-score = -2.73, R) >droYak2.chr2R 10099787 99 + 21139217 CUCACUGUAAGGACACGCUGUACUCUGACCUUACAGCUG------AACAUGUUCGCGACGCCCUAUUAUCACAGCCGUGCGCACAUGUAU------UUGUUUACUUGGACG .(((((((((((.((..........)).)))))))).))------)((((((.(((.(((..((........)).)))))).))))))..------..((((....)))). ( -25.00, z-score = -1.14, R) >droSec1.super_1 7670266 99 + 14215200 CUCACUGUAAGGACACGCUGUGCUCUGACCUUACAGCUG------AACAUGUGCGCGACGCCCUAUUAUCACAGCCGUGCGCACAUGUAU------UUGUUUACUUGGACG .(((((((((((.((.((...))..)).)))))))).))------)((((((((((.(((..((........)).)))))))))))))..------..((((....)))). ( -32.90, z-score = -2.73, R) >droSim1.chr2R 8925506 99 + 19596830 CUCACUGUAAGGACACGCUGUGCUCUGACCUUACAGCUU------AACAUGUGCGCGACGCCCUAUUAUCACAGCCGUGCGCACAUGUAU------UUGUUUACUUGGACG ....((((((((.((.((...))..)).))))))))...------.((((((((((.(((..((........)).)))))))))))))..------..((((....)))). ( -30.50, z-score = -2.26, R) >droMoj3.scaffold_6496 12808570 93 + 26866924 UGCACUGUAAGGACGCGCUCUGCUCUGACCUUACAGCUACAGCUGAACAUGUGCGCGCCGCGCGC------------UAUGCACAUGUAU------UUGUUUACUUGGCCG ....((((((((.((.((...))..)).)))))))).....((..(((((((((((((...))))------------...))))))))..------........)..)).. ( -28.00, z-score = 0.10, R) >droVir3.scaffold_12875 19119555 77 + 20611582 CACACUGUAAGGACGCGUUCUGCUCUGACCUU--------------ACAUGUGCACGCCGUCC--------------CAUGCACAUGUAU------UUGUUUACUUGGCCG ..((..(((((((.((.....))))).....(--------------(((((((((.(......--------------).)))))))))).------....)))).)).... ( -17.40, z-score = -0.10, R) >droPer1.super_31 555606 110 - 935084 CUUAUUGUAAGGACACGCUCUGUUCUU-CUCUAUCCUUACGGUCGAACAUGUGCACGCUGCCACAUUAUAACAGCCGCGCGCACAUGUACGGGUAGUUGUUUACUCGGACG .....((((((((..............-.....))))))))(((..((((((((.(((.((............)).))).)))))))).(((((((....)))))))))). ( -37.01, z-score = -3.19, R) >dp4.chr3 17412850 110 + 19779522 CUUAUUGUAAGGACACGCUCUGUUCUU-CUCUAUCCUUACGGUCGAACAUGUGCACGCUGCCACAUUAUAACAGCCGCGCGCACAUGUACGGGUAGUUGUUUACUCGGACG .....((((((((..............-.....))))))))(((..((((((((.(((.((............)).))).)))))))).(((((((....)))))))))). ( -37.01, z-score = -3.19, R) >consensus CUCACUGUAAGGACACGCUGUGCUCUGACCUUACAGCUG______AACAUGUGCGCGACGCCCUAUUAUCACAGCCGUGCGCACAUGUAU______UUGUUUACUUGGACG ......((((((.((.((...))..)).))))))............(((((((((((..((............))..)))))))))))....................... (-18.23 = -18.59 + 0.36)
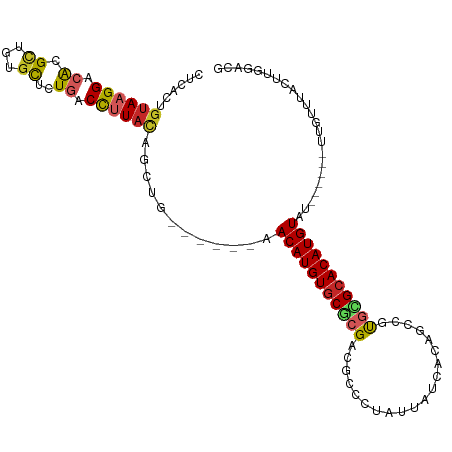
| Location | 10,162,610 – 10,162,709 |
|---|---|
| Length | 99 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.05 |
| Shannon entropy | 0.46087 |
| G+C content | 0.50990 |
| Mean single sequence MFE | -36.33 |
| Consensus MFE | -15.53 |
| Energy contribution | -15.04 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.979101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
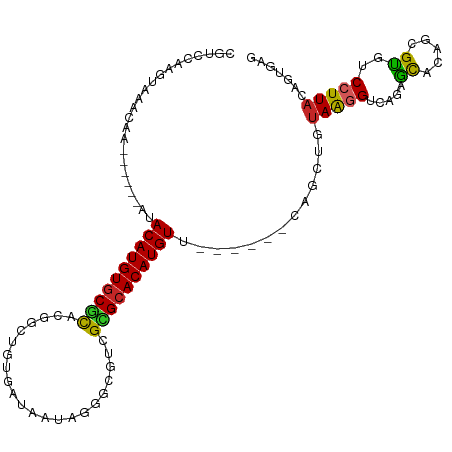
>dm3.chr2R 10162610 99 - 21146708 CGUCCAAGUAAACAA------AUACAUGUGCGCACGGCUGUGAUAAUAGGGCGUCGCGCACAUGUU------CAGCUGUAAGGUCAGAGCACAGCGUGUCCUUACAGUGAG ...............------..(((((((((((((.((.........)).))).))))))))))(------((.((((((((.((..((...)).)).))))))))))). ( -38.60, z-score = -3.33, R) >droAna3.scaffold_13266 14512666 105 - 19884421 CGUCCAAGUAAACAA------AUACAUGUGCGCACGACUGUGAUAAUAGGACGUCGCGCACAUGUUCAUGUUCAGCUGUAAGGUCAGAGCUCAGCGUGUCCUUACAGUGAG ..........((((.------..(((((((((((((.((((....))))..))).))))))))))...))))..(((((((((.((..((...)).)).)))))))))... ( -41.50, z-score = -4.27, R) >droEre2.scaffold_4845 6909901 99 + 22589142 CGUCCAAGUAAACAA------AUACAUGUGCGCACGGCUGUGAUAAUAGGGCGUCGCGCACAUGUU------CAGCUGUAAGGUCAGAGCACAGCGUGUCCUUACAGUGAG ...............------..(((((((((((((.((.........)).))).))))))))))(------((.((((((((.((..((...)).)).))))))))))). ( -38.60, z-score = -3.33, R) >droYak2.chr2R 10099787 99 - 21139217 CGUCCAAGUAAACAA------AUACAUGUGCGCACGGCUGUGAUAAUAGGGCGUCGCGAACAUGUU------CAGCUGUAAGGUCAGAGUACAGCGUGUCCUUACAGUGAG ...............------..((((((.((((((.((.........)).))).))).))))))(------((.((((((((.((..(.....).)).))))))))))). ( -30.90, z-score = -1.51, R) >droSec1.super_1 7670266 99 - 14215200 CGUCCAAGUAAACAA------AUACAUGUGCGCACGGCUGUGAUAAUAGGGCGUCGCGCACAUGUU------CAGCUGUAAGGUCAGAGCACAGCGUGUCCUUACAGUGAG ...............------..(((((((((((((.((.........)).))).))))))))))(------((.((((((((.((..((...)).)).))))))))))). ( -38.60, z-score = -3.33, R) >droSim1.chr2R 8925506 99 - 19596830 CGUCCAAGUAAACAA------AUACAUGUGCGCACGGCUGUGAUAAUAGGGCGUCGCGCACAUGUU------AAGCUGUAAGGUCAGAGCACAGCGUGUCCUUACAGUGAG ...............------..(((((((((((((.((.........)).))).)))))))))).------..(((((((((.((..((...)).)).)))))))))... ( -38.20, z-score = -3.37, R) >droMoj3.scaffold_6496 12808570 93 - 26866924 CGGCCAAGUAAACAA------AUACAUGUGCAUA------------GCGCGCGGCGCGCACAUGUUCAGCUGUAGCUGUAAGGUCAGAGCAGAGCGCGUCCUUACAGUGCA ..((.......(((.------..((((((((...------------((((...)))))))))))).....))).(((((((((.....((.....))..))))))))))). ( -32.00, z-score = -0.33, R) >droVir3.scaffold_12875 19119555 77 - 20611582 CGGCCAAGUAAACAA------AUACAUGUGCAUG--------------GGACGGCGUGCACAUGU--------------AAGGUCAGAGCAGAACGCGUCCUUACAGUGUG .((((..(....)..------.((((((((((((--------------......)))))))))))--------------).))))........((((.........)))). ( -25.20, z-score = -1.76, R) >droPer1.super_31 555606 110 + 935084 CGUCCGAGUAAACAACUACCCGUACAUGUGCGCGCGGCUGUUAUAAUGUGGCAGCGUGCACAUGUUCGACCGUAAGGAUAGAG-AAGAACAGAGCGUGUCCUUACAAUAAG .(((((.(((......))).)).((((((((((((.((..(......)..)).))))))))))))..))).(((((((((...-............)))))))))...... ( -39.86, z-score = -3.51, R) >dp4.chr3 17412850 110 - 19779522 CGUCCGAGUAAACAACUACCCGUACAUGUGCGCGCGGCUGUUAUAAUGUGGCAGCGUGCACAUGUUCGACCGUAAGGAUAGAG-AAGAACAGAGCGUGUCCUUACAAUAAG .(((((.(((......))).)).((((((((((((.((..(......)..)).))))))))))))..))).(((((((((...-............)))))))))...... ( -39.86, z-score = -3.51, R) >consensus CGUCCAAGUAAACAA______AUACAUGUGCGCACGGCUGUGAUAAUAGGGCGUCGCGCACAUGUU______CAGCUGUAAGGUCAGAGCACAGCGUGUCCUUACAGUGAG .......................((((((((((......................)))))))))).............(((((.....((.....))..)))))....... (-15.53 = -15.04 + -0.49)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:22:11 2011