| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,134,570 – 10,134,660 |
| Length | 90 |
| Max. P | 0.618689 |

| Location | 10,134,570 – 10,134,660 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 66.34 |
| Shannon entropy | 0.63846 |
| G+C content | 0.48437 |
| Mean single sequence MFE | -27.27 |
| Consensus MFE | -8.99 |
| Energy contribution | -8.78 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.618689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
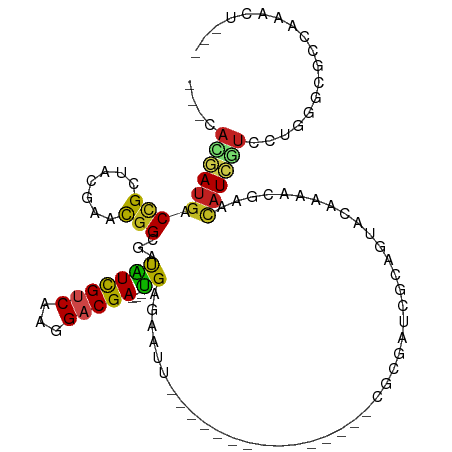
>dm3.chr2R 10134570 90 + 21146708 ---CACGAUGACCGUUAAGAACGGCGAUGUCGUCAAGGACGA---UGUGAAUU---------------CGCGAUCGCAGUACAAAACGAACAUCGUCCUGGGCGCCAAACU--- ---.....((.((((.....)))))).((.((((.(((((((---(((.....---------------.((....)).((.....))..)))))))))).)))).))....--- ( -28.70, z-score = -1.41, R) >droSim1.chr2R 8897083 90 + 19596830 ---CACGAUGACCGUUAAGAACGGCGAUAUCGUCAAGGACGA---UGUGAAUU---------------CGCGAUCGCAGUACAAAACGAACAUCGUCCUGGGCGCCAAACU--- ---...((((.((((.....))))...))))(((.(((((((---(((.....---------------.((....)).((.....))..)))))))))).)))........--- ( -27.50, z-score = -1.53, R) >droSec1.super_1 7642124 90 + 14215200 ---CACGAUGACCGUUAAGAACGGCGAUAUCGUCAAGGACGA---UGUGAAUU---------------CGCGAUCGCAGUACAAAACGAACAUCGUCCUGGGCGUCAAACU--- ---...((((.((.........((((....)))).(((((((---(((.....---------------.((....)).((.....))..)))))))))).)))))).....--- ( -29.50, z-score = -2.09, R) >droYak2.chr2R 10071395 90 + 21139217 ---CACAAUGACCGUUAAGAACGGCGAUGUCGUCAAGGACGA---CGGGAAUU---------------CGCGAUCGCAGUACAAAACGAACAUCGUCCUGGGCGCCAAACU--- ---.(((.((.((((.....)))))).)))((((.(((((((---((.(....---------------).)).(((..........)))...))))))).)))).......--- ( -24.60, z-score = -0.34, R) >droEre2.scaffold_4845 6879900 90 - 22589142 ---CACGAUGACCGUUAAGAACGGCGAUAUCGUCAAGGACGA---UGUGGAUU---------------CGCGGUCGCAGUACAAAACGAACAUCGUCCUGGGCGCCAAACU--- ---...((((.((((.....))))...))))(((.(((((((---(((.....---------------.((....)).((.....))..)))))))))).)))........--- ( -27.50, z-score = -0.84, R) >droAna3.scaffold_13266 14484637 90 + 19884421 ---UAUAAUGACUGCAACAAAUGGCGUUAUCGCCAAGGACGA---UGAAAAAU---------------CGCGGCAACAUUACAAAACAAACAUUGUCCUUGGACAACAAUU--- ---....(((((.((........))))))).(((((((((((---((......---------------...(....).((....))....)))))))))))).).......--- ( -21.50, z-score = -3.04, R) >dp4.chr3 17385257 96 + 19779522 ---AAGGAUGACCGCCUCGAACGGCGUCAUCGUCAAGGACGACGAUGACAAAU---------------CGCGUUUGCAGUACAAAACGAACAUCGUCCUGGGAUCAGCAGCAUU ---.(((((((..(((......)))(((((((((......)))))))))....---------------..(((((........)))))....)))))))............... ( -32.30, z-score = -2.09, R) >droPer1.super_31 528028 96 - 935084 ---AAGGAUGACAGCCUCGAACGGCGUCAUCGUCAAGGACGACGAUGACAAAU---------------CGCGUUUGCAGUACAAAACGAACAUCGUCCUGGGAUCAGCAGCAUU ---...((((((.(((......)))))))))(((.(((((((((((.....))---------------))(((((........)))))....)))))))..))).......... ( -32.30, z-score = -2.47, R) >droWil1.scaffold_180697 3457220 93 - 4168966 CACAAUGACUGCCGCAUCGAACGGUAACAUCGUCCAGGACGA---UGAGGGAC---------------CACCGUCCCAUUAUAAAGCCAAUAUAACAUUGGGUGACUCCUU--- ..(((((..(((((.......))))).(((((((...)))))---)).(((((---------------....)))))..................)))))(....).....--- ( -27.30, z-score = -2.12, R) >droVir3.scaffold_12875 19089213 93 + 20611582 UCUGAUAAUGACCGCGGUGAACGGCAACGUCGCCAAGGACGA---CGAAAAUC---------------GUUUGUCACAUUAUAAAACGAAUAUUGUCUUGGGCGAAUCCUU--- ............((((((((.((....)))))))......((---(((.(.((---------------((((...........)))))).).)))))....))).......--- ( -25.00, z-score = -1.16, R) >droMoj3.scaffold_6496 12769789 96 + 26866924 CAUCAAAAUGACCGAGUCUAACGGCAAUGUCGUCAAGGACGA---CGAUAGCA---------AUC---GAUUGUCACGCUAUGCAACGAAUAUUGUUUUGGCUGACUCCUU--- .............(((((....((((((((((((...)))))---)(((....---------)))---.))))))..((((.((((......))))..)))).)))))...--- ( -26.40, z-score = -1.68, R) >droGri2.scaffold_15245 13118852 108 + 18325388 UAAAAUAAUGCCCGCGUCAAACGGAAAUGUUGUCAAGGACGA---CGACAAUCCCGGCUUAUAUCCCAAGUUAUCCCAAUAUAAAUCGAAUAUUGUUUUGGGCGAUUCAUU--- ........((((((...(((..(((..(((((((......))---))))).)))((..((((((..............))))))..))....)))...)))))).......--- ( -24.64, z-score = -0.92, R) >consensus ___CACGAUGACCGCUACGAACGGCGAUAUCGUCAAGGACGA___UGAGAAUU_______________CGCGAUCGCAGUACAAAACGAACAUCGUCCUGGGCGCCAAACU___ ...........((.........((((....)))).(((((((...((.......................................))....)))))))))............. ( -8.99 = -8.78 + -0.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:22:10 2011