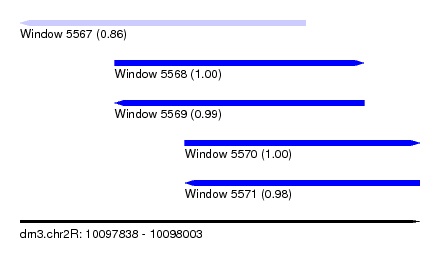
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,097,838 – 10,098,003 |
| Length | 165 |
| Max. P | 0.999686 |

| Location | 10,097,838 – 10,097,956 |
|---|---|
| Length | 118 |
| Sequences | 9 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 78.33 |
| Shannon entropy | 0.44849 |
| G+C content | 0.38026 |
| Mean single sequence MFE | -26.23 |
| Consensus MFE | -11.44 |
| Energy contribution | -11.14 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.61 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
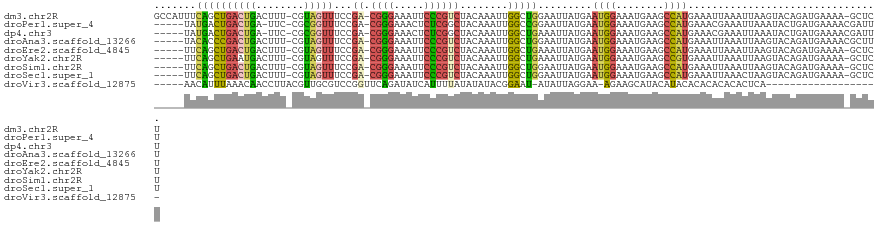
>dm3.chr2R 10097838 118 - 21146708 GCCAUUUCAGCUGACUGACUUU-CGUAGUUUCCGA-CGGGAAAUUCCCGUCUACAAAUUGGCUGGAAUUAUGAAUGGAAAUGAAGCCAUGAAAUUAAAUUAAGUACAGAUGAAAA-GCUCU ((..(((((.(((....((((.-..(((((((.((-((((.....)))))).......(((((...(((.........)))..))))).)))))))....)))).))).))))).-))... ( -32.20, z-score = -2.80, R) >droPer1.super_4 6718496 113 + 7162766 -----UAUGACUGACUGA-UUC-CGCGGUUUCCGA-CGGGAAACUCUCGGCUACAAAUUGGCCGGAAUUAUGAAUGGAAAUGAAGCCAUGAAACGAAAUUAAAUACUGAUGAAAACGAUUU -----..........(((-(((-(..(((((((..-..)))))))...(((((.....))))))))))))...((((........))))................................ ( -24.40, z-score = -1.72, R) >dp4.chr3 16320272 113 - 19779522 -----UAUGACUGACUGA-UUC-CGCGGUUUCCGA-CGGGAAACUCUCGGCUACAAAUUGGCUGAAAUUAUGAAUGGAAAUGAAGCCAUGAAACGAAAUUAAAUACUGAUGAAAACGAUUU -----((((.((......-(((-((.(((((((..-..))))))).(((((((.....))))))).........)))))....)).))))............................... ( -21.20, z-score = -1.43, R) >droAna3.scaffold_13266 14059369 114 + 19884421 -----UACACCCGACUGACUUU-CGUAGUUUCCGA-CGGGAAAUUCCCGUCUACAAAUUGGCUGGAAUUAUGAAUGGAAAUGAAGCCAUGAAAUUAAAUUAAGUACAGAUGAAAACGCUUU -----......((.(((((((.-..(((((((.((-((((.....)))))).......(((((...(((.........)))..))))).)))))))....)))).))).)).......... ( -28.50, z-score = -2.67, R) >droEre2.scaffold_4845 6844363 113 + 22589142 -----UUCAGCUGACUGACUUU-CGUAGUUUCCGA-CGGGAAAUUCCCGUCUACAAAUUGGCUGAAAUUAUGAAUGGAAAUGAAGCCAUGAAAUUAAAUUAAGUACAGAUGAAAA-GCUCU -----...((((..(((((((.-..(((((((.((-((((.....)))))).......(((((...(((.........)))..))))).)))))))....)))).)))......)-))).. ( -30.40, z-score = -3.24, R) >droYak2.chr2R 10035529 113 - 21139217 -----UUCAGCUGAAUGACUUU-CGUAGUUUCCGA-CGGGAAAUUCCCGUCUACAAAUUGGCUGAAAUUAUGAAUGGAAAUGAAGCCGUGAAAUUAAAUUAAGUACAGAUGAAAA-GCUCU -----((((.(((....((((.-..((((((((((-((((.....))))))........((((...(((.........)))..))))).)))))))....)))).))).))))..-..... ( -29.90, z-score = -2.97, R) >droSim1.chr2R 8861118 113 - 19596830 -----UUCAGCUGACUGACUUU-CGUAGUUUCCGA-CGGGAAAUUCCCGUCUACAAAUUGGCUGGAAUUAUGAAUGGAAAUGAAGCCAUGAAAUUAAAUUAAGUACAGAUGAAAA-GCUCU -----...((((..(((((((.-..(((((((.((-((((.....)))))).......(((((...(((.........)))..))))).)))))))....)))).)))......)-))).. ( -30.40, z-score = -2.81, R) >droSec1.super_1 7606156 113 - 14215200 -----UUCAGCUGACUGACUUU-CGUAGUUUCCGA-CGGGAAAUUCCCGUCUACAAAUUGGCUGGAAUUAUGAAUGGAAAUGAAGCCAUGAAAUUAAACUAAGUACAGAUGAAAA-GCUCU -----...((((..(((((((.-..(((((((.((-((((.....)))))).......(((((...(((.........)))..))))).)))))))....)))).)))......)-))).. ( -30.40, z-score = -2.84, R) >droVir3.scaffold_12875 2991325 95 + 20611582 -----AACAUUUAAACAACCUUACGUUGCGUCCGGUUCAGAUAUCAUUUUAUAUAUACGGAAU-AUAUUAGGAA-AGAAGCAUACAUACACACACACACUCA------------------- -----...................(((((.((...(((.(((((.(((((........)))))-))))).))).-.)).))).)).................------------------- ( -8.70, z-score = 0.10, R) >consensus _____UUCAGCUGACUGACUUU_CGUAGUUUCCGA_CGGGAAAUUCCCGUCUACAAAUUGGCUGGAAUUAUGAAUGGAAAUGAAGCCAUGAAAUUAAAUUAAGUACAGAUGAAAA_GCUCU .......(((((((............(((((((.....)))))))............))))))).........((((........))))................................ (-11.44 = -11.14 + -0.30)
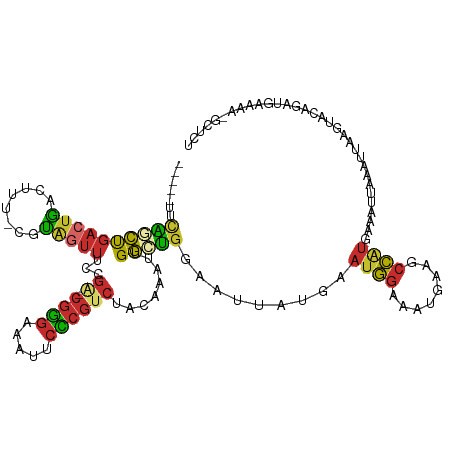
| Location | 10,097,877 – 10,097,980 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 85.55 |
| Shannon entropy | 0.28303 |
| G+C content | 0.40595 |
| Mean single sequence MFE | -30.87 |
| Consensus MFE | -21.22 |
| Energy contribution | -22.42 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.71 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.30 |
| SVM RNA-class probability | 0.998257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10097877 103 + 21146708 AUUUCCAUUCAUAAUUCCAGCCAAUUUGUAGACGGGAAUUUCCCGUCGGAAACUACGAAAGUCAGUCAGCUGAAAUGGCUAGCUAAUACGUAUAAAGGGGAAA .(..((............(((((.((((((((((((.....))))))(....)))))))..((((....))))..))))).((......)).....))..).. ( -31.60, z-score = -3.40, R) >droAna3.scaffold_13266 14059409 90 - 19884421 AUUUCCAUUCAUAAUUCCAGCCAAUUUGUAGACGGGAAUUUCCCGUCGGAAACUACGAAAGUCAGUCGGGUGUA--CCUCAGUUACUAUAUA----------- .....(((((....((((.((......)).((((((.....))))))))))(((((....)).))).)))))..--................----------- ( -22.30, z-score = -1.94, R) >droEre2.scaffold_4845 6844402 94 - 22589142 AUUUCCAUUCAUAAUUUCAGCCAAUUUGUAGACGGGAAUUUCCCGUCGGAAACUACGAAAGUCAGUCAGCUGAA-UGGCUAA--------UAUAUAGGGGAAG ..((((.(..(((..((.(((((.((((((((((((.....))))))(....)))))))..((((....)))).-)))))))--------..)))..))))). ( -30.90, z-score = -4.23, R) >droYak2.chr2R 10035568 102 + 21139217 AUUUCCAUUCAUAAUUUCAGCCAAUUUGUAGACGGGAAUUUCCCGUCGGAAACUACGAAAGUCAUUCAGCUGAA-UGUCUAAUACAUAUGUAUAUAGGGGAAG ..((((.........((((((.........((((((.....)))))).(((...((....))..))).))))))-..((((((((....)))).)))))))). ( -30.20, z-score = -3.88, R) >droSim1.chr2R 8861157 100 + 19596830 AUUUCCAUUCAUAAUUCCAGCCAAUUUGUAGACGGGAAUUUCCCGUCGGAAACUACGAAAGUCAGUCAGCUGAA-UGGCUAAU--ACGUAUAUAUGGGGGAAG .(..((((..(((.....(((((.((((((((((((.....))))))(....)))))))..((((....)))).-)))))...--...)))..))))..)... ( -35.10, z-score = -4.40, R) >droSec1.super_1 7606195 100 + 14215200 AUUUCCAUUCAUAAUUCCAGCCAAUUUGUAGACGGGAAUUUCCCGUCGGAAACUACGAAAGUCAGUCAGCUGAA-UGGCUAAU--ACGUAUAUAUGGGGGAAG .(..((((..(((.....(((((.((((((((((((.....))))))(....)))))))..((((....)))).-)))))...--...)))..))))..)... ( -35.10, z-score = -4.40, R) >consensus AUUUCCAUUCAUAAUUCCAGCCAAUUUGUAGACGGGAAUUUCCCGUCGGAAACUACGAAAGUCAGUCAGCUGAA_UGGCUAAU__AUAUAUAUAUAGGGGAAG ..................(((((.((((((((((((.....))))))(....)))))))..((((....))))..)))))....................... (-21.22 = -22.42 + 1.20)

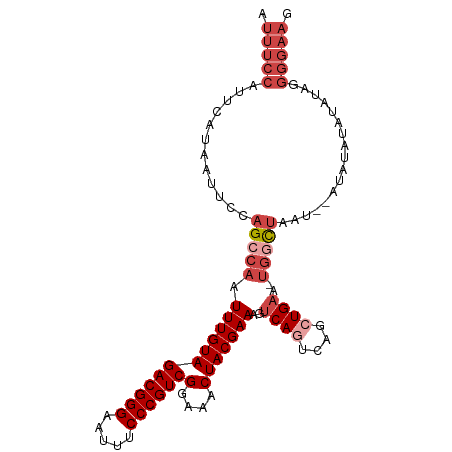
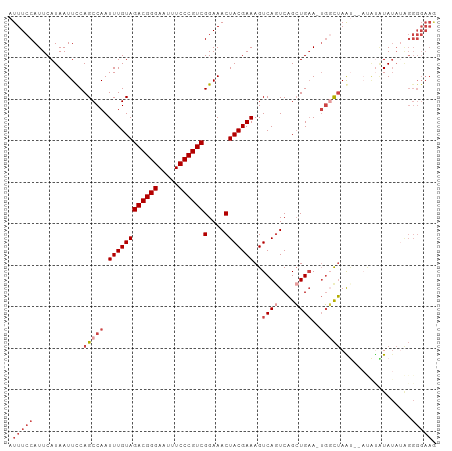
| Location | 10,097,877 – 10,097,980 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 85.55 |
| Shannon entropy | 0.28303 |
| G+C content | 0.40595 |
| Mean single sequence MFE | -27.33 |
| Consensus MFE | -21.65 |
| Energy contribution | -22.27 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.993858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10097877 103 - 21146708 UUUCCCCUUUAUACGUAUUAGCUAGCCAUUUCAGCUGACUGACUUUCGUAGUUUCCGACGGGAAAUUCCCGUCUACAAAUUGGCUGGAAUUAUGAAUGGAAAU .......(((((.((((....(((((((..((((....))))..............((((((.....)))))).......)))))))...)))).)))))... ( -29.30, z-score = -2.91, R) >droAna3.scaffold_13266 14059409 90 + 19884421 -----------UAUAUAGUAACUGAGG--UACACCCGACUGACUUUCGUAGUUUCCGACGGGAAAUUCCCGUCUACAAAUUGGCUGGAAUUAUGAAUGGAAAU -----------....((((...((.((--....)))))))).(.(((((((((((.((((((.....))))))..(.....)...))))))))))).)..... ( -21.80, z-score = -1.12, R) >droEre2.scaffold_4845 6844402 94 + 22589142 CUUCCCCUAUAUA--------UUAGCCA-UUCAGCUGACUGACUUUCGUAGUUUCCGACGGGAAAUUCCCGUCUACAAAUUGGCUGAAAUUAUGAAUGGAAAU .((((..((((..--------(((((((-.((((....))))..............((((((.....)))))).......)))))))...))))...)))).. ( -27.20, z-score = -3.69, R) >droYak2.chr2R 10035568 102 - 21139217 CUUCCCCUAUAUACAUAUGUAUUAGACA-UUCAGCUGAAUGACUUUCGUAGUUUCCGACGGGAAAUUCCCGUCUACAAAUUGGCUGAAAUUAUGAAUGGAAAU .((((.(((.((((....))))))).((-(((....)))))...(((((((((((.((((((.....))))))..(.....)...))))))))))).)))).. ( -29.50, z-score = -3.90, R) >droSim1.chr2R 8861157 100 - 19596830 CUUCCCCCAUAUAUACGU--AUUAGCCA-UUCAGCUGACUGACUUUCGUAGUUUCCGACGGGAAAUUCCCGUCUACAAAUUGGCUGGAAUUAUGAAUGGAAAU ......((((.((((.((--.(((((((-.((((....))))..............((((((.....)))))).......))))))).)))))).)))).... ( -28.10, z-score = -2.78, R) >droSec1.super_1 7606195 100 - 14215200 CUUCCCCCAUAUAUACGU--AUUAGCCA-UUCAGCUGACUGACUUUCGUAGUUUCCGACGGGAAAUUCCCGUCUACAAAUUGGCUGGAAUUAUGAAUGGAAAU ......((((.((((.((--.(((((((-.((((....))))..............((((((.....)))))).......))))))).)))))).)))).... ( -28.10, z-score = -2.78, R) >consensus CUUCCCCUAUAUAUAUAU__AUUAGCCA_UUCAGCUGACUGACUUUCGUAGUUUCCGACGGGAAAUUCCCGUCUACAAAUUGGCUGGAAUUAUGAAUGGAAAU .((((................(((((.......)))))......(((((((((((.((((((.....))))))..(.....)...))))))))))).)))).. (-21.65 = -22.27 + 0.61)

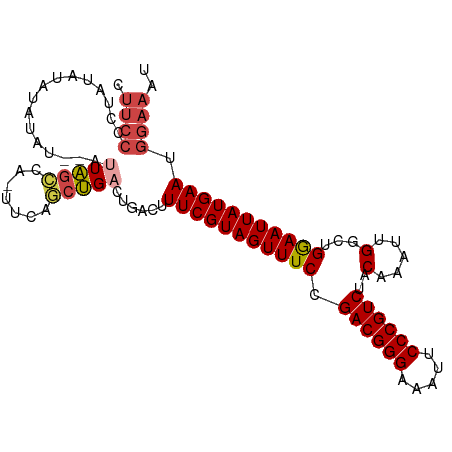
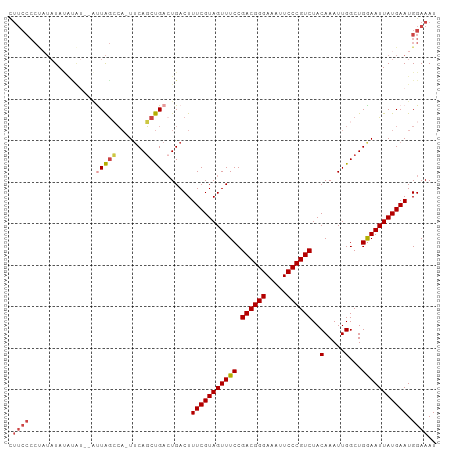
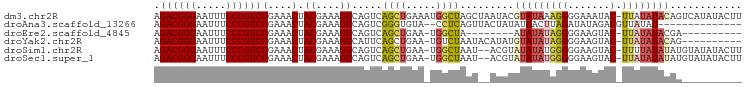
| Location | 10,097,906 – 10,098,003 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 75.60 |
| Shannon entropy | 0.45047 |
| G+C content | 0.39449 |
| Mean single sequence MFE | -28.42 |
| Consensus MFE | -17.76 |
| Energy contribution | -18.43 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.27 |
| Mean z-score | -3.77 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.19 |
| SVM RNA-class probability | 0.999686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10097906 97 + 21146708 AGACGGGAAUUUCCCGUCGGAAACUACGAAAGUCAGUCAGCUGAAAUGGCUAGCUAAUACGUAUAAAGGGGAAAUAC-UUAUAUACAGUCAUAUACUU .((((((.....))))))(....).((....)).((..((((.....))))..)).....(((((.(((.......)-)).)))))............ ( -26.60, z-score = -2.99, R) >droAna3.scaffold_13266 14059438 82 - 19884421 AGACGGGAAUUUCCCGUCGGAAACUACGAAAGUCAGUCGGGUGUA--CCUCAGUUACUAUAUAACUUAGAUAUAGACGUUAUAU-------------- .((((((.....))))))....(((((....)).)))((..((((--.((.(((((.....))))).)).))))..))......-------------- ( -23.20, z-score = -1.95, R) >droEre2.scaffold_4845 6844431 78 - 22589142 AGACGGGAAUUUCCCGUCGGAAACUACGAAAGUCAGUCAGCUGAA-UGGCUA--------AUAUAUAGGGGAAGUAC-UUAUAUACGA---------- .((((((.....))))))(....)..((..(((((.((....)).-))))).--------.((((((((.......)-))))))))).---------- ( -25.50, z-score = -3.59, R) >droYak2.chr2R 10035597 86 + 21139217 AGACGGGAAUUUCCCGUCGGAAACUACGAAAGUCAUUCAGCUGAA-UGUCUAAUACAUAUGUAUAUAGGGGAAGUAC-UUAUAUACAG---------- .((((((.....))))))(....)......((.(((((....)))-)).))........((((((((((.......)-))))))))).---------- ( -29.50, z-score = -4.86, R) >droSim1.chr2R 8861186 94 + 19596830 AGACGGGAAUUUCCCGUCGGAAACUACGAAAGUCAGUCAGCUGAA-UGGCUAAU--ACGUAUAUAUGGGGGAAGUAC-UUUUAUAUAUGUAUAUACUU .((((((.....))))))(....).((....)).(((((......-))))).((--(((((((((..((.......)-)..)))))))))))...... ( -31.20, z-score = -4.09, R) >droSec1.super_1 7606224 94 + 14215200 AGACGGGAAUUUCCCGUCGGAAACUACGAAAGUCAGUCAGCUGAA-UGGCUAAU--ACGUAUAUAUGGGGGAAGUAC-UUAUAUAUAUGUAUAUACUU .((((((.....))))))(....).((....)).(((((......-))))).((--(((((((((((((.......)-))))))))))))))...... ( -34.50, z-score = -5.15, R) >consensus AGACGGGAAUUUCCCGUCGGAAACUACGAAAGUCAGUCAGCUGAA_UGGCUAAU__AUAUAUAUAUAGGGGAAGUAC_UUAUAUACAG__________ .((((((.....))))))(....).((....)).....((((.....)))).........((((((((.(......).))))))))............ (-17.76 = -18.43 + 0.67)
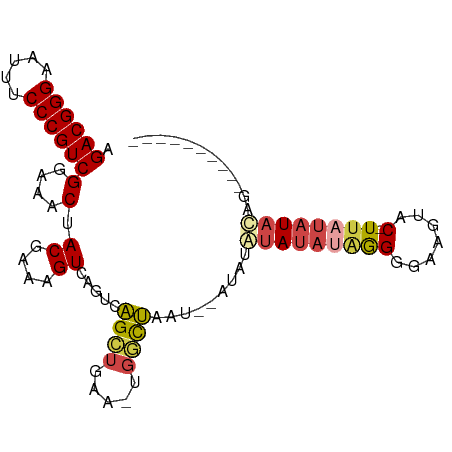
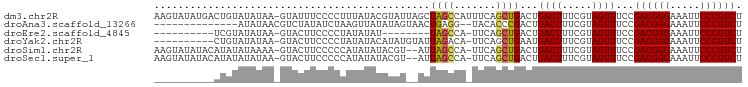
| Location | 10,097,906 – 10,098,003 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 75.60 |
| Shannon entropy | 0.45047 |
| G+C content | 0.39449 |
| Mean single sequence MFE | -20.17 |
| Consensus MFE | -13.05 |
| Energy contribution | -12.88 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.984655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10097906 97 - 21146708 AAGUAUAUGACUGUAUAUAA-GUAUUUCCCCUUUAUACGUAUUAGCUAGCCAUUUCAGCUGACUGACUUUCGUAGUUUCCGACGGGAAAUUCCCGUCU .....(((((..(((((.((-(........))))))))...((((.((((.......)))).))))...)))))......((((((.....)))))). ( -22.60, z-score = -2.22, R) >droAna3.scaffold_13266 14059438 82 + 19884421 --------------AUAUAACGUCUAUAUCUAAGUUAUAUAGUAACUGAGG--UACACCCGACUGACUUUCGUAGUUUCCGACGGGAAAUUCCCGUCU --------------......((..(.(((((.((((((...)))))).)))--)).)..))...((((.....))))...((((((.....)))))). ( -20.20, z-score = -1.40, R) >droEre2.scaffold_4845 6844431 78 + 22589142 ----------UCGUAUAUAA-GUACUUCCCCUAUAUAU--------UAGCCA-UUCAGCUGACUGACUUUCGUAGUUUCCGACGGGAAAUUCCCGUCU ----------..((((....-))))............(--------((((..-....)))))..((((.....))))...((((((.....)))))). ( -17.20, z-score = -1.96, R) >droYak2.chr2R 10035597 86 - 21139217 ----------CUGUAUAUAA-GUACUUCCCCUAUAUACAUAUGUAUUAGACA-UUCAGCUGAAUGACUUUCGUAGUUUCCGACGGGAAAUUCCCGUCU ----------.((((((((.-(.......).))))))))((((....((.((-(((....))))).))..))))......((((((.....)))))). ( -22.60, z-score = -3.21, R) >droSim1.chr2R 8861186 94 - 19596830 AAGUAUAUACAUAUAUAAAA-GUACUUCCCCCAUAUAUACGU--AUUAGCCA-UUCAGCUGACUGACUUUCGUAGUUUCCGACGGGAAAUUCCCGUCU ..(((((((...........-............)))))))..--.(((((..-....)))))..((((.....))))...((((((.....)))))). ( -18.80, z-score = -1.89, R) >droSec1.super_1 7606224 94 - 14215200 AAGUAUAUACAUAUAUAUAA-GUACUUCCCCCAUAUAUACGU--AUUAGCCA-UUCAGCUGACUGACUUUCGUAGUUUCCGACGGGAAAUUCCCGUCU ((((...(((.(((((((..-...........))))))).))--)(((((..-....)))))...))))...........((((((.....)))))). ( -19.62, z-score = -1.99, R) >consensus __________AUGUAUAUAA_GUACUUCCCCUAUAUAUAUAU__AUUAGCCA_UUCAGCUGACUGACUUUCGUAGUUUCCGACGGGAAAUUCCCGUCU ..............................................((((.......))))...((((.....))))...((((((.....)))))). (-13.05 = -12.88 + -0.16)
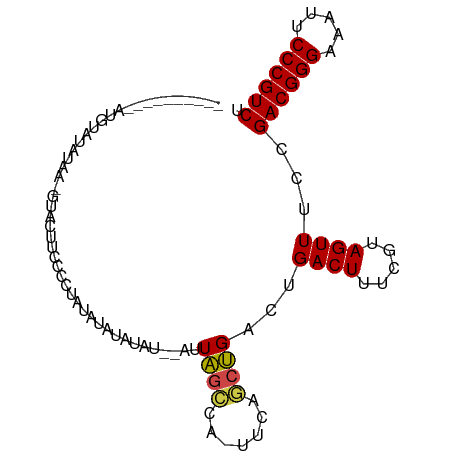
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:22:04 2011