
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,094,263 – 10,094,363 |
| Length | 100 |
| Max. P | 0.898017 |

| Location | 10,094,263 – 10,094,363 |
|---|---|
| Length | 100 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 68.02 |
| Shannon entropy | 0.53340 |
| G+C content | 0.49355 |
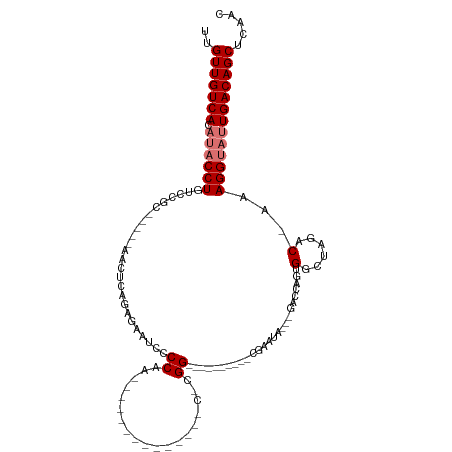
| Mean single sequence MFE | -26.76 |
| Consensus MFE | -10.66 |
| Energy contribution | -11.42 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.898017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
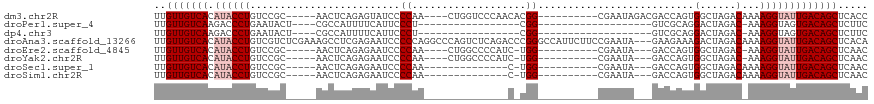
>dm3.chr2R 10094263 100 - 21146708 UUGUUGUCACAUACCUGUCCGC-----AACUCAGAGUAUCCCCAA----CUGGUCCCAACACGG----------CGAAUAGACGACCAGUGGCUAGACAAAAGGUAUUGACAGCUCACC ..(((((((.(((((((((.((-----........))....(((.----((((((((.....))----------.........)))))))))...)))...)))))))))))))..... ( -27.40, z-score = -1.58, R) >droPer1.super_4 6715697 78 + 7162766 UUGUUGUCAAGACCCUGAAUACU----CGCCAUUUUCAUUCCCU-----------------CGG-------------------GUCGCAGGACUAGAC-AAAGGUAGUGACAGCUCUUC ..(((((((..((((((...(((----((...............-----------------)))-------------------))..)))).((....-..))))..)))))))..... ( -18.76, z-score = -0.44, R) >dp4.chr3 16317455 78 - 19779522 UUGUUGUCAAGACCCUGAAUACU----CGCCAUUUUCAUUCCCU-----------------CGG-------------------GUCGCAGGACUAGAC-AAAGGUAGUGACAGCUCUUC ..(((((((..((((((...(((----((...............-----------------)))-------------------))..)))).((....-..))))..)))))))..... ( -18.76, z-score = -0.44, R) >droAna3.scaffold_13266 14055860 116 + 19884421 UUGUUGUCACAUACCUGUCGUCUCGAAAGCCUCGAGAAUCCCCCAGGCCCAGUCUCAGACCCGGGCCAUUCUUCCGAAUA---GAAGAAAGACUAGACAAAAGGUAUUGACAGCUCACA ..(((((((.(((((((((.((((((.....))))))........(((((.(((...)))..))))).((((((......---))))))......)))...)))))))))))))..... ( -42.40, z-score = -5.14, R) >droEre2.scaffold_4845 6840554 95 + 22589142 UUGUUGUCACAUACCUGUCCGC-----AACUCAGAGAAUCCCCAA----CUGGCCCCAUC-UGG----------CGAAUA---GACCAGUGGCUAGAC-AAAGGUAUUGACAGCUCAAC ..(((((((.((((((.(((..-----......).))........----((((((....(-(((----------(.....---).)))).))))))..-..)))))))))))))..... ( -29.70, z-score = -2.68, R) >droYak2.chr2R 10031458 95 - 21139217 UUGUUGUCACAUACCUGUCCGC-----AACUCAGAGAAUCCCCAA----CUGGCCCCAUC-UGG----------CGAAUA---GACCAGUGGCUAGAC-AAAGGUAUUGACAGCUCAAC ..(((((((.((((((.(((..-----......).))........----((((((....(-(((----------(.....---).)))).))))))..-..)))))))))))))..... ( -29.70, z-score = -2.68, R) >droSec1.super_1 7602210 86 - 14215200 UUGUUGUCACAUACCUGUCCGC-----AACUCAGAGAAUCCCCAA--------------C-UGG----------CGAAUA---GACCAGUGGCUAGACAAAAGGUAUUGACAGCUCAAC ..(((((((.((((((((((..-----......).......(((.--------------(-(((----------(.....---).)))))))...)))...)))))))))))))..... ( -23.70, z-score = -2.09, R) >droSim1.chr2R 8857205 86 - 19596830 UUGUUGUCACAUACCUGUCCGC-----AACUCAGAGAAUCCCCAA--------------C-UGG----------CGAAUA---GACCAGUGGCUAGACAAAAGGUAUUGACAGCUCAAC ..(((((((.((((((((((..-----......).......(((.--------------(-(((----------(.....---).)))))))...)))...)))))))))))))..... ( -23.70, z-score = -2.09, R) >consensus UUGUUGUCACAUACCUGUCCGC_____AACUCAGAGAAUCCCCAA______________C_CGG__________CGAAUA___GACCAGUGGCUAGAC_AAAGGUAUUGACAGCUCAAC ..(((((((.((((((.....................................................................................)))))))))))))..... (-10.66 = -11.42 + 0.75)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:22:01 2011