
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,090,688 – 10,090,752 |
| Length | 64 |
| Max. P | 0.566607 |

| Location | 10,090,688 – 10,090,752 |
|---|---|
| Length | 64 |
| Sequences | 14 |
| Columns | 79 |
| Reading direction | forward |
| Mean pairwise identity | 63.19 |
| Shannon entropy | 0.73675 |
| G+C content | 0.48695 |
| Mean single sequence MFE | -16.58 |
| Consensus MFE | -7.25 |
| Energy contribution | -7.25 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.59 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.566607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10090688 64 + 21146708 UGCGAUCUUGCGGCGUCAGCGAUCGCCAUUUUUUUCCUGCAGC--GAAAAGAUGAAGAUAAAGUGU------------- .((.(((((((((((....)).))))(((((((((.(....).--))))))))))))))...))..------------- ( -15.50, z-score = 0.13, R) >droSim1.chr2R 8853527 64 + 19596830 UGCGAUCUUGUGGCGUUAGCGAUCGCCAUUUUUUUCCUGAAGC--GAGAAGAUGAAGAUAAAGUGU------------- .((.(((((((((((........))))))((((((((....).--)))))))..)))))...))..------------- ( -15.50, z-score = -0.86, R) >droSec1.super_1 7598594 64 + 14215200 UGCGAUCUUGUGGCGUCAGCGAUCGCCAUUUUUUUCCUGCAGU--GAAAAGAUGAAGAUGAAGUGU------------- .(((((((((......))).))))))((((((((..(....).--.))))))))............------------- ( -15.80, z-score = -0.60, R) >droYak2.chr2R 10027734 64 + 21139217 UGCGAUCUUGGGGCGUUAGUGACCGCCACUUCUUUCCUGCAGC--GAAAAAAAUCAGAAUGAAUUU------------- (((((....((((((........)))).))....))..)))..--.....................------------- ( -9.20, z-score = 1.56, R) >droEre2.scaffold_4845 6836982 64 - 22589142 UGCGAUCUUGCGGCGUUAGCGAACGCCAUUUCUUUCCUGCAGC--GAGAAGAUUUAGAUAGAAUGU------------- .....((((((((((((....))))))....(......)..))--)))).................------------- ( -14.80, z-score = 0.19, R) >droAna3.scaffold_13266 14051972 61 - 19884421 UUCGAUCUUGGGGCGUCAGGGAACGCCAUUUUUUGCCUGAAA---AAUAUAAAGAAAAUGGUGU--------------- .....((((((....)))))).((((((((((((........---.......))))))))))))--------------- ( -14.56, z-score = -1.16, R) >dp4.chr3 16313858 64 + 19779522 UGCGGUCCUGCGGCGUCAAUGAGCGCCAUUUCUUGCCUGGAAC--GAGAUUAGGAGAAGAGAUUAC------------- .(((....)))(((((......)))))(((((((.(((((...--....)))))..)))))))...------------- ( -17.90, z-score = -0.62, R) >droPer1.super_4 6712116 64 - 7162766 UGCGGUCCUGCGGCGUCAAUGAGCGCCAUUUCUUGCCUGGAAC--GAGAUGAGGAGAAGAGAUUAC------------- .(((....)))(((((......)))))(((((((.(((.(...--....).)))..)))))))...------------- ( -16.80, z-score = -0.09, R) >droWil1.scaffold_180700 4741508 74 - 6630534 GUCGAUCUUGGGGCGUCAAUGAACGCCAUUUCUUUCCUGGAA---AUUCA--CAAGAGUCAGAAGUCAGAAAAAAGUUU ...(((((((.(((((......)))))((((((.....))))---))...--))))).))................... ( -16.70, z-score = -0.65, R) >droVir3.scaffold_12875 2982281 66 - 20611582 GACGAUCCUGUGGCGUCAAGGAGCGCCACUUUUUGCCUGGAAGGCGCAAACAGUGGCAUUAGAUGG------------- ((((.((....)))))).......((((((.(((((((....)).))))).)))))).........------------- ( -21.60, z-score = -0.95, R) >droMoj3.scaffold_6496 15614844 70 - 26866924 GACGAUCCUGCGGCGUCAGGGAGCGCCACUUUUUGCCUG-----CAGAAAAACAGAUAUUAGACCCUGGAGCUGG---- ..........((((..(((((.(((........)))(((-----........)))........)))))..)))).---- ( -19.00, z-score = -0.25, R) >droGri2.scaffold_15245 14054242 59 + 18325388 GCCGAUCCUGCGGCGUUAAGGAGCGCCACUUUUUGCCUG-------UUAACAGGAGCAUUAGUUGG------------- .(((((..(((((((((....))))))........((((-------....)))).)))...)))))------------- ( -21.00, z-score = -1.92, R) >anoGam1.chr3L 36064891 63 + 41284009 GACGAUCCUGGGGCGUUAGAGAGCGCCACUUUUUACCUGGUG---AAGAGAGGAAGAAUACAUUAG------------- .....((((..((((((....)))))).((((((((...)))---)))))))))............------------- ( -18.60, z-score = -2.53, R) >triCas2.ChLG7 8153043 65 - 17478683 GUCUGUCUUGCGGCGUCAAAGAGCGCCACUUCUUGCCUGCAA---ACGAAGACAAAAAUUACAAGUGU----------- ((((...((((((.(.(((..((.....))..))))))))))---....))))...............----------- ( -15.20, z-score = -0.48, R) >consensus UGCGAUCUUGCGGCGUCAGCGAGCGCCAUUUUUUGCCUGCAA___GAGAAGAGGAAAAUAAAAUGU_____________ ...........((((........)))).................................................... ( -7.25 = -7.25 + 0.00)

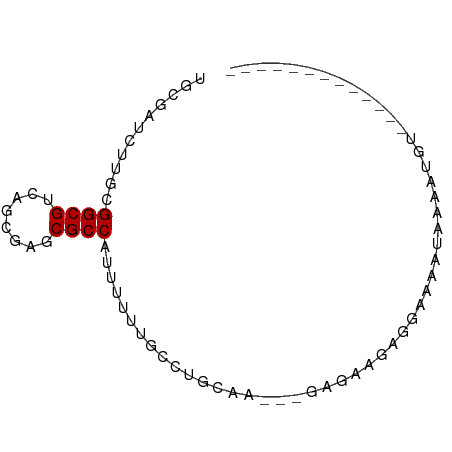
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:22:00 2011