
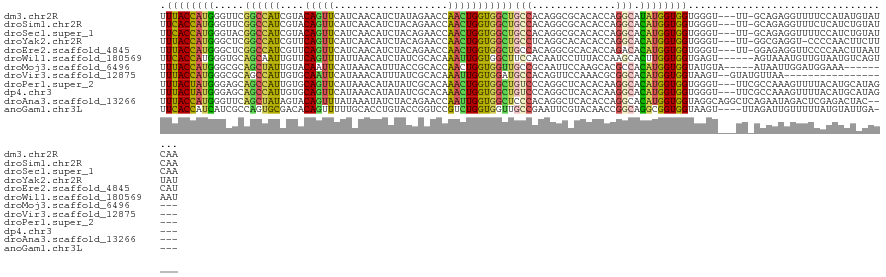
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,045,554 – 10,045,673 |
| Length | 119 |
| Max. P | 0.597705 |

| Location | 10,045,554 – 10,045,673 |
|---|---|
| Length | 119 |
| Sequences | 12 |
| Columns | 123 |
| Reading direction | forward |
| Mean pairwise identity | 68.42 |
| Shannon entropy | 0.70231 |
| G+C content | 0.47935 |
| Mean single sequence MFE | -33.81 |
| Consensus MFE | -15.50 |
| Energy contribution | -14.79 |
| Covariance contribution | -0.71 |
| Combinations/Pair | 1.68 |
| Mean z-score | -0.64 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.597705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
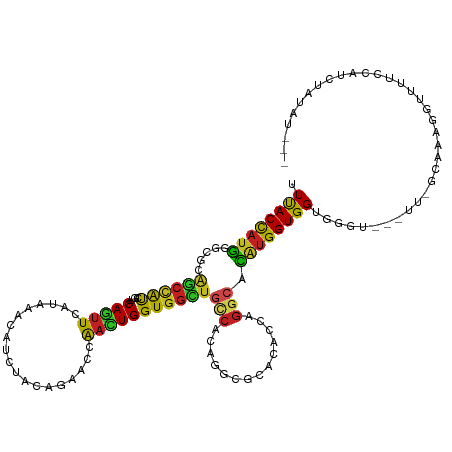
>dm3.chr2R 10045554 119 + 21146708 UUUACCAUGGGUUCGGCCAUCGUACAGUUCAUCAACAUCUAUAGAACCAACUGGUGGCUGCCACAGGCGCACACCAGGCAUAUGGUGGUGGGU---UU-GCAGAGGUUUUCCAUAUGUAUCAA ..(((.(((((...((((.(((((.((..((((....((....))((((.((((((((.(((...))))).)))))).....))))))))..)---))-)).)))))).)))))..))).... ( -35.30, z-score = -0.49, R) >droSim1.chr2R 8814928 119 + 19596830 UUCACCAUGGGUUCGGCCAUCGUACAGUUCAUCAACAUCUACAGAACCAACUGGUGGCUGCCACAGGCGCACACCAGGCACAUGGUGGUGGGU---UU-GCAGAGGUUUCUCAUCUGUAUCAA .(((((((((((((.((....))...(((....))).......)))))..((((((((.(((...))))).))))))...)))))))).....---.(-(((((.(.....).)))))).... ( -38.90, z-score = -1.32, R) >droSec1.super_1 7553226 119 + 14215200 UUCACCAUGGGUACGGCCAUCGUACAGUUCAUCAACAUCUACAGAACCAACUGGUGGCUGCCACAGGCGCACACCAGGCACAUGGUGGUGGGU---UU-GCAGAGGUUUUCCAUCUGUAUCAA .((((((((.(((((.....))))).((((.............))))...((((((((.(((...))))).))))))...)))))))).....---.(-(((((((....)).)))))).... ( -42.12, z-score = -2.02, R) >droYak2.chr2R 9981785 118 + 21139217 UUUACCAUGGGCUCGGCCAUCGUUCAGUUCAUCAACAUCUACAGAACCAACUGGUGGCUGCCUCAGGCACACACCAGGCACAUGGUGGUGGGU---UU-GGCGAGGU-CCCCAACUUCUUUAU .......((((...((((.((((..((..((((....((....))((((.((((((..((((...))))..)))))).....))))))))..)---).-.)))))))-))))).......... ( -41.50, z-score = -1.85, R) >droEre2.scaffold_4845 6796727 119 - 22589142 UUUACCAUGGGCUCGGCCAUCGUUCAGUUCAUCAACAUCUACAGAACCAACUGGUGGCUGCCACAGGCGCACACCAGACACAUGGUGGUGGGU---UU-GGAGAGGUUCCCCAACUUAAUCAU .......(((((((((((((((((..((((.............)))).))).)))))))....(((((.(((((((......)))).))).))---))-)..)))....)))).......... ( -35.82, z-score = -0.33, R) >droWil1.scaffold_180569 375607 117 - 1405142 UUCACCAUGGGUGCAGCAAUUGUUCAGUUUAUUAACAUCUAUCGCACAAAUUGGUGGCUUCCACAAUCCUUUACCAAGCACUUGGUGGUGAGU------AGUAAAUGUUGUAAUGUCAGUAAU .((((((...(((((((....)))..(((....))).......))))....)))))).....(((((..(((((.(..((((....))))..)------.))))).)))))............ ( -24.00, z-score = 0.66, R) >droMoj3.scaffold_6496 13615220 109 + 26866924 UUUACCAUGGGCGCAGCUAUUGUACAAUUCAUAAACAUUUACCGCACCAACUGGUGGUUGCCGCAAUUCCAAGCACGCCACAUGGUGGUAUGUA-----AUAAUUGGAUGGAAA--------- .((((((((((((((((...(((...........))).......((((....))))))))).((........))..))).))))))))......-----...............--------- ( -27.60, z-score = 0.44, R) >droVir3.scaffold_12875 18736099 102 - 20611582 UUUACCAUGGGCGCAGCCAUUGUGCAAUUCAUAAACAUUUAUCGCACAAAUUGGUGGAUGCCACAGUUCCAAACGCGGCACAUGGUGGUAAGU--GUAUGUUAA------------------- .(((((((((.(((.((((((((((..................))))))..))))(((.........)))....))).).)))))))).....--.........------------------- ( -29.17, z-score = -0.47, R) >droPer1.super_2 2933135 117 + 9036312 UUUACUAUGGGAGCAGCCAUUGUGCAGUUCAUAAACAUAUAUCGCACAAACUGGUGGCUGUCCCAGGCUCACACAAGGCACAUGGUGGUGGGU---UUCGCCAAAGUUUUACAUGCAUAG--- .......(((((.((((((((((((.((............)).))))).....)))))))))))).((......(((((...((((((.....---.))))))..)))))....))....--- ( -33.30, z-score = -0.47, R) >dp4.chr3 2743126 117 + 19779522 UUUACUAUGGGAGCAGCCAUUGUGCAGUUCAUAAACAUAUAUCGCACAAACUGGUGGCUGUCCCAGGCUCACACAAGGCACAUGGUGGUGGGU---UUCGCCAAAGUUUUACAUGCAUAG--- .......(((((.((((((((((((.((............)).))))).....)))))))))))).((......(((((...((((((.....---.))))))..)))))....))....--- ( -33.30, z-score = -0.47, R) >droAna3.scaffold_13266 14009997 118 - 19884421 UUUACCAUGGGUUCAGCUAUAGUACAGUUUAUAAAUAUCUACAGAACCAAUUGGUGGCUCCCACAGGCUCACACCAGGCACAUGGUGGUAGGCAGGCUCAGAAUAGACUCGAGACUAC----- ..((((((((((((.((....))...((..((....))..)).)))))..(((((((..((....))..).))))))...)))))))((((.....(((((......)).))).))))----- ( -27.30, z-score = 0.40, R) >anoGam1.chr3L 8221594 115 - 41284009 UUCACCAUCAUCGCCAGUGCGACACAGUUUUUGCACCUGUACCGGUCCGUCUGGUGGUUGCCGAAUUCGUACAACCGGCAGGCGGUGGUAAGU----UUAGAUUGUUUUUAUGUAUUGA---- ..............(((((((..(((((((..((.....(((((..(((((((.((((((.((....))..)))))).)))))))))))).))----..))))))).....))))))).---- ( -37.40, z-score = -1.80, R) >consensus UUUACCAUGGGCGCAGCCAUCGUACAGUUCAUAAACAUCUACAGAACCAACUGGUGGCUGCCACAGGCGCACACCAGGCACAUGGUGGUGGGU___UU_GCAAAGGUUUUCCAUCUAUAU___ .((((((((.....((((((....(((((...................)))))))))))(((..............))).))))))))................................... (-15.50 = -14.79 + -0.71)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:21:56 2011