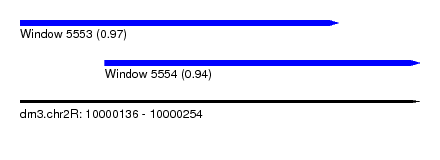
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 10,000,136 – 10,000,254 |
| Length | 118 |
| Max. P | 0.971546 |

| Location | 10,000,136 – 10,000,230 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 62.05 |
| Shannon entropy | 0.82478 |
| G+C content | 0.49405 |
| Mean single sequence MFE | -29.72 |
| Consensus MFE | -13.38 |
| Energy contribution | -13.63 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.971546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
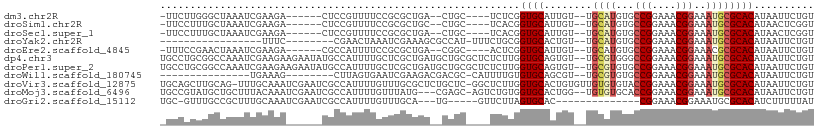
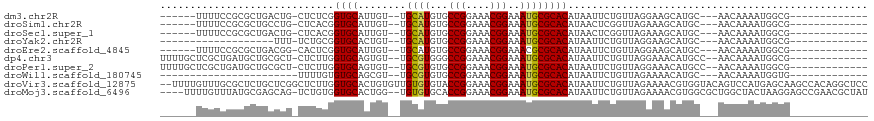
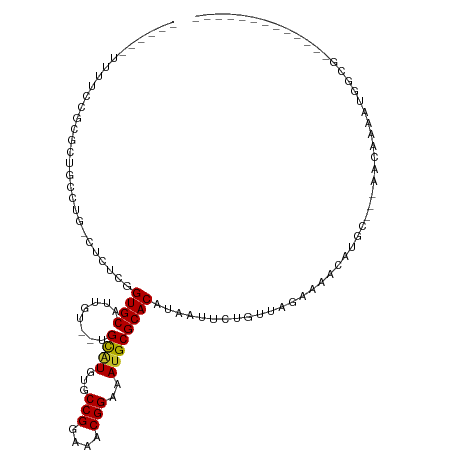
>dm3.chr2R 10000136 94 + 21146708 -UUCUUGGGCUAAAUCGAAGA------CUCCGUUUUCCGCGCUGA--CUGC----UCUCGGUGCAUUGU--UGCAUGUGCCGGAAACGGAAAUGCGCACAUAAUUCUGU -....((.((......(((((------(...)))))).(((((((--....----..))))))).....--.((((...(((....)))..)))))).))......... ( -27.70, z-score = -1.03, R) >droSim1.chr2R 8433939 94 + 19596830 -UUCCUUUGCUAAAUCGAAGA------CUCCGUUUUCCGCGCUGC--CUGC----UCACGGUGCAUUGU--UGCAUGUGCCGGAAACGGAAAUGCGCACAUAACUCGGU -...(((((......))))).------..(((......((((((.--....----...))))))..(((--(((((...(((....)))..))))).))).....))). ( -27.80, z-score = -0.86, R) >droSec1.super_1 7508214 94 + 14215200 -UUCCUUUGCUAAAUCGAAGA------CUCCGUUUUCCGCGCUGA--CUGC----UCACGGUGCAUUGU--UGCAUGUGCCGGAAACGGAAAUGCGCACAUAACUCGGU -...(((((......))))).------..(((......((((((.--....----...))))))..(((--(((((...(((....)))..))))).))).....))). ( -27.80, z-score = -1.08, R) >droYak2.chr2R 9932896 81 + 21139217 -----------------UUUC--------CGAACUAAAUCGAAAGCGCCAU-UUUCUGCGGUGCACUGU--UGCAUGUGCCGGAAACGGAAAUGCGCACAUAAUUCUGU -----------------....--------(((......)))...(((((..-.......)))))..(((--(((((...(((....)))..))))).)))......... ( -23.40, z-score = -1.40, R) >droEre2.scaffold_4845 6751435 94 - 22589142 -UUUCCGAACUAAAUCGAAGA------CGCCAUUUUCCGCGCUGA--CGGC----ACUCGGUGCAUUGU--UGCAUGUGCCGGAAACGGAAACGCGCACAUAAUUCUGU -.....(((.......(((((------.....))))).((((...--((((----((...(((((....--))))))))))).....(....)))))......)))... ( -27.20, z-score = -0.64, R) >dp4.chr3 2694525 107 + 19779522 UGCCUGCGGCCAAAUCGAAGAAGAAUAUGCCAUUUUGCUCGCUGAUGCUGCGCUCUCUUGGUGCAGUGU--UGCGUGGGCCGGAAACGGAAAUGCGCACAUAAUUCUGU .(((...)))........((((..((.(((((((((((((((((((((((((((.....))))))))))--)).))))))((....)))))))).))).))..)))).. ( -39.30, z-score = -1.70, R) >droPer1.super_2 2883944 107 + 9036312 UGCCUGCGGCCAAAUCGAAGAAGAAUAUGCCAUUUUGCUCGCUGAUGCUGCGCUCUCUUGGUGCAGUGU--UGCGUGUGCCGGAAACGGAAAUGCGCACAUAAUUCUGU .(((...)))........((((..((.((((((((.((.(((.(((((((((((.....))))))))))--)))).)).(((....)))))))).))).))..)))).. ( -37.30, z-score = -1.49, R) >droWil1.scaffold_180745 1239186 83 - 2843958 ---------------UGAAAG--------CUUAGUGAAUCGAAGACGACGC-CAUUUUGUGUGCAGCGU--UGCGUGUGCCGGAAACGGAAAUGCGCACAUAAUUCUGU ---------------.(((..--------....(((..(((....))))))-....((((((((.((((--(.......(((....))).))))))))))))))))... ( -23.20, z-score = -0.26, R) >droVir3.scaffold_12875 18681763 107 - 20611582 UGCAGCUUGCAG-UUUGCAAAUCGAAUCGCCAUUUUGUUUGCGCUCUGCUC-GGCUCUUGGUGCACUGUGUUGUGUGUACCGGAAACGGAAAUGCGCACAUAAUUCUGU ...((((.((((-..(((((((.((((....)))).)))))))..))))..-))))......(((..((..(((((((((((....)))...))))))))..))..))) ( -35.30, z-score = -1.33, R) >droMoj3.scaffold_6496 23336294 103 + 26866924 UGCCGUAUGCUGCUUUACAAAUCGAAUCGCCAUUUUGUUUAUG---CGAGC-AGUCUGUGGUGCACUGG--UGUGUGCACCGGAAACGGAAAUGCGCACAUAAUUCUGU .((((((.(((((((.......((((.......))))......---.))))-))).))))))(((...(--(((((((((((....)))...))))))))).....))) ( -34.74, z-score = -1.62, R) >droGri2.scaffold_15112 1138122 86 - 5172618 UGC-GUUUGCCGCUUUGCAAAUCGAAUCGCCAUUUUGUUUGCA---UG-----GUUCUUAGUGCAC--------------CGGAAACGGAAAUGCGCACAUCUUUUUAU (((-((.(((((((.(((((((.((((....)))).)))))))---.)-----)).....).)))(--------------((....)))....)))))........... ( -23.20, z-score = -0.77, R) >consensus _GCCCUUUGCUAAAUCGAAGA______CGCCAUUUUGCUCGCUGA__CUGC____UCUCGGUGCACUGU__UGCAUGUGCCGGAAACGGAAAUGCGCACAUAAUUCUGU ............................................................((((........((((...(((....)))..)))))))).......... (-13.38 = -13.63 + 0.25)
| Location | 10,000,161 – 10,000,254 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 69.37 |
| Shannon entropy | 0.58360 |
| G+C content | 0.49543 |
| Mean single sequence MFE | -32.84 |
| Consensus MFE | -15.00 |
| Energy contribution | -14.74 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.942090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 10000161 93 + 21146708 ------UUUUCCGCGCUGACUG-CUCUCGGUGCAUUGU--UGCAUGUGCCGGAAACGGAAAUGCGCACAUAAUUCUGUUAGGAAGCAUGC---AACAAAAUGGCG------------- ------....(((((((((...-...))))))).((((--(((((((.(((....)))..............((((....))))))))))---)))))...))..------------- ( -34.80, z-score = -2.83, R) >droSim1.chr2R 8433964 93 + 19596830 ------UUUUCCGCGCUGCCUG-CUCACGGUGCAUUGU--UGCAUGUGCCGGAAACGGAAAUGCGCACAUAACUCGGUUAGAAAGCAUGC---AACAAAAUGGCG------------- ------....((((((((....-....)))))).((((--(((((((.(((....)))...............((.....))..))))))---)))))...))..------------- ( -31.30, z-score = -1.15, R) >droSec1.super_1 7508239 93 + 14215200 ------UUUUCCGCGCUGACUG-CUCACGGUGCAUUGU--UGCAUGUGCCGGAAACGGAAAUGCGCACAUAACUCGGUUAGAAAGCAUGC---AACAAAAUGGCG------------- ------....((((((((....-....)))))).((((--(((((((.(((....)))...............((.....))..))))))---)))))...))..------------- ( -31.30, z-score = -1.60, R) >droYak2.chr2R 9932921 80 + 21139217 -------------------UUU-UCUGCGGUGCACUGU--UGCAUGUGCCGGAAACGGAAAUGCGCACAUAAUUCUGUUAGGAAGCAUGC---AACAAAAUGGCG------------- -------------------...-...((.((....(((--(((((((.(((....)))..............((((....))))))))))---))))..)).)).------------- ( -26.60, z-score = -1.91, R) >droEre2.scaffold_4845 6751460 93 - 22589142 ------UUUUCCGCGCUGACGG-CACUCGGUGCAUUGU--UGCAUGUGCCGGAAACGGAAACGCGCACAUAAUUCUGUUAGGAAGCAUGC---AACAAAAUGGCG------------- ------....(((((((((...-...))))))).((((--(((((((.(((....)))..............((((....))))))))))---)))))...))..------------- ( -34.80, z-score = -2.44, R) >dp4.chr3 2694557 100 + 19779522 UUUUGCUCGCUGAUGCUGCGCU-CUCUUGGUGCAGUGU--UGCGUGGGCCGGAAACGGAAAUGCGCACAUAAUUCUGUUAGGAAACAUGCC--AACAAAAUGGCG------------- .(((.((.((.(((((((((((-.....))))))))))--)(((((..(((....)))...)))))..........)).)).)))...(((--(......)))).------------- ( -35.60, z-score = -1.90, R) >droPer1.super_2 2883976 100 + 9036312 UUUUGCUCGCUGAUGCUGCGCU-CUCUUGGUGCAGUGU--UGCGUGUGCCGGAAACGGAAAUGCGCACAUAAUUCUGUUAGGAAACAUGCC--AACAAAAUGGCG------------- ...(((.....(((((((((((-.....))))))))))--)((((...(((....)))..))))))).............(....)..(((--(......)))).------------- ( -35.60, z-score = -2.06, R) >droWil1.scaffold_180745 1239216 77 - 2843958 -----------------------UUUUGUGUGCAGCGU--UGCGUGUGCCGGAAACGGAAAUGCGCACAUAAUUCUGUUAGAAAACAUGC---AACAAAAUGGUG------------- -----------------------((((((.((((..((--(((((((.(((....)))..))))))......(((.....)))))).)))---))))))).....------------- ( -23.50, z-score = -1.38, R) >droVir3.scaffold_12875 18681794 116 - 20611582 --UUUUGUUUGCGCUCUGCUCGGCUCUUGGUGCACUGUGUUGUGUGUACCGGAAACGGAAAUGCGCACAUAAUUCUGUUAGAAAACGUGGUACAGUCCAUGAGCAAGCCACAGGCUCC --....(((((.(((.((((((.....(((((.((..((((((((((((((....)))...)))))))....(((.....)))))))..)).))..))))))))))))..)))))... ( -37.70, z-score = -1.06, R) >droMoj3.scaffold_6496 23336326 111 + 26866924 ----UUUUGUUUAUGCGAGCAG-UCUGUGGUGCACUGG--UGUGUGCACCGGAAACGGAAAUGCGCACAUAAUUCUGUUAGAAAACGUGGCGCUGGCUACUAAGGAGCCGAACGCUAU ----....((((.((((.((..-...))..))))...(--(((((((((((....)))...)))))))))............))))((((((.(((((.(....))))))..)))))) ( -37.20, z-score = -1.32, R) >consensus ______UUUUCCGCGCUGCCUG_CUCUCGGUGCAUUGU__UGCAUGUGCCGGAAACGGAAAUGCGCACAUAAUUCUGUUAGAAAACAUGC___AACAAAAUGGCG_____________ .............................((((........((((...(((....)))..)))))))).................................................. (-15.00 = -14.74 + -0.26)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:21:50 2011