
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,988,068 – 9,988,126 |
| Length | 58 |
| Max. P | 0.652544 |

| Location | 9,988,068 – 9,988,126 |
|---|---|
| Length | 58 |
| Sequences | 4 |
| Columns | 58 |
| Reading direction | forward |
| Mean pairwise identity | 62.93 |
| Shannon entropy | 0.62247 |
| G+C content | 0.37069 |
| Mean single sequence MFE | -11.79 |
| Consensus MFE | -7.01 |
| Energy contribution | -6.33 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.71 |
| Mean z-score | -0.47 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.652544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
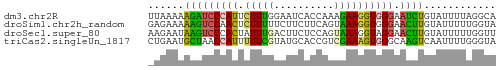
>dm3.chr2R 9988068 58 + 21146708 UUAAAAAGAUCCCAUUCUUUGGAAUCACCAAAGAAGGUGGGAAUCUGUAUUUUAGGCA ((((((((((((((((((((((.....)))))))..))))).))))...))))))... ( -18.40, z-score = -3.02, R) >droSim1.chr2h_random 793290 58 + 3178526 GAGAAAAAGUCCAACUCUUUUUCUUCUUCAGUAAAGGUGGGAACUUGUAUUUUUGGUA (((((((((.......)))))))))....(.((((((((.(....).)))))))).). ( -9.00, z-score = 0.55, R) >droSec1.super_80 44630 58 + 135508 AAGAAUAAGUCCCACUAUUUGACUUCUCCAGUAAAGGUAGGAACUUGUAUUUUUGGUU ....((((((((.(((.....(((.....)))...))).)).)))))).......... ( -8.40, z-score = 0.66, R) >triCas2.singleUn_1817 15298 58 - 22356 CUGAAUGCUAACCAUUUUUCGUAUGCACCGUCGAAAGUGGGCAAGUCAAUUUUGGGUA ...........(((.........(((.(((.(....))))))).........)))... ( -11.37, z-score = -0.05, R) >consensus AAGAAAAAGUCCCACUCUUUGUAUUCACCAGUAAAGGUGGGAACUUGUAUUUUUGGUA ......(((((((((.(((((..........)))))))))).))))............ ( -7.01 = -6.33 + -0.69)
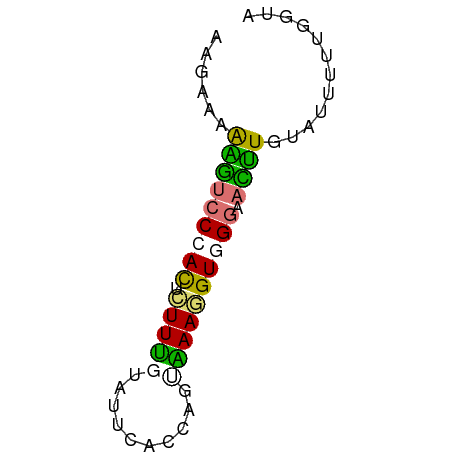
| Location | 9,988,068 – 9,988,126 |
|---|---|
| Length | 58 |
| Sequences | 4 |
| Columns | 58 |
| Reading direction | reverse |
| Mean pairwise identity | 62.93 |
| Shannon entropy | 0.62247 |
| G+C content | 0.37069 |
| Mean single sequence MFE | -10.15 |
| Consensus MFE | -5.32 |
| Energy contribution | -4.45 |
| Covariance contribution | -0.87 |
| Combinations/Pair | 1.64 |
| Mean z-score | -0.72 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.628508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
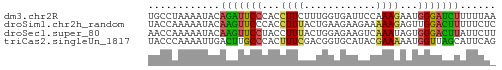
>dm3.chr2R 9988068 58 - 21146708 UGCCUAAAAUACAGAUUCCCACCUUCUUUGGUGAUUCCAAAGAAUGGGAUCUUUUUAA ....(((((...((((.((((..((((((((.....))))))))))))))))))))). ( -17.80, z-score = -3.62, R) >droSim1.chr2h_random 793290 58 - 3178526 UACCAAAAAUACAAGUUCCCACCUUUACUGAAGAAGAAAAAGAGUUGGACUUUUUCUC ............((((.((.((((((.((.....))..)))).)).))))))...... ( -6.20, z-score = 0.59, R) >droSec1.super_80 44630 58 - 135508 AACCAAAAAUACAAGUUCCUACCUUUACUGGAGAAGUCAAAUAGUGGGACUUAUUCUU ............((((.(((((..(((((.....)))..))..)))))))))...... ( -9.30, z-score = -0.10, R) >triCas2.singleUn_1817 15298 58 + 22356 UACCCAAAAUUGACUUGCCCACUUUCGACGGUGCAUACGAAAAAUGGUUAGCAUUCAG ..........(((..((((((.(((((..........)))))..)))...))).))). ( -7.30, z-score = 0.26, R) >consensus UACCAAAAAUACAAGUUCCCACCUUCACUGGAGAAGACAAAGAAUGGGACUUAUUCAA ............(((((((...((((............))))...)))))))...... ( -5.32 = -4.45 + -0.87)
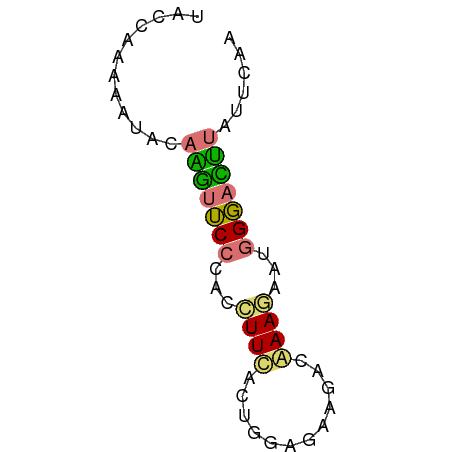
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:21:48 2011