| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,971,697 – 9,971,800 |
| Length | 103 |
| Max. P | 0.880721 |

| Location | 9,971,697 – 9,971,800 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 65.01 |
| Shannon entropy | 0.65284 |
| G+C content | 0.29190 |
| Mean single sequence MFE | -20.74 |
| Consensus MFE | -9.26 |
| Energy contribution | -8.98 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.880721 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
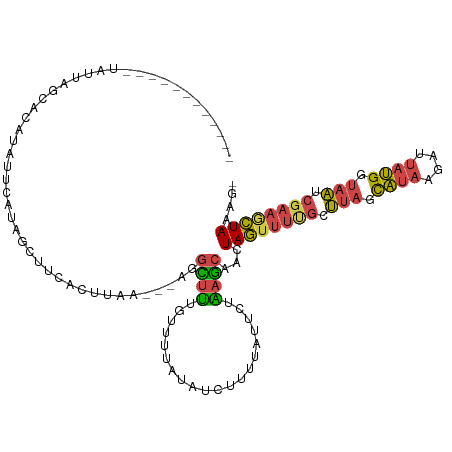
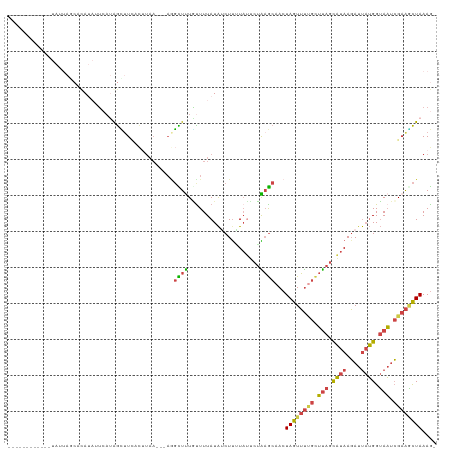
>dm3.chr2R 9971697 103 + 21146708 ------------UAUUACCACAUAUUCAUGGUUUAACUUAA---AUGUUUUGUUUUAUAUCUUGUAUUCUAAGCAACUAGUUUUGCUUAGCAUAAGAUUAUGGUAAUCGAAGCUAAAG- ------------....((((........)))).........---..((((((..(((((((((((...((((((((......)))))))).)))))).)))))....)))))).....- ( -20.70, z-score = -1.94, R) >droWil1.scaffold_180745 1224073 107 - 2843958 ------------UAUUAUAAUAUUUACUAAAGUUGAUCUAAGACAACUUCUUUUUUAUGCCUUUUUAACAGCACAACUAAUGCAAGACAGUUUAAUAUCCAGGAAUCAGCGUUUAAAAG ------------.................((((((........))))))(((((..((((..........(((.......)))..((...(((........))).)).))))..))))) ( -9.60, z-score = 1.13, R) >droEre2.scaffold_4845 6739147 115 - 22589142 AAUAUGCCAGUGUAUUAGCACAGAUUCAUAGCAUCACUUAA---AGGCUCUGUUUUAUAUCUCAUAUUCUGAGCAACUAGUUUUGUUUAGCAUAAGAUUAUGGUAAUCGAAACUAAAG- ....((((((.((((.((....((..((.(((.........---..))).))..))....)).)))).))).)))..((((((((.(((.((((....)))).))).))))))))...- ( -23.80, z-score = -1.14, R) >droYak2.chr2R 9920430 109 + 21139217 AAAAUGACAGUGUACUAGCACAUAUUCAUGGUUUUA---------GGUUUUACUUUAUAUCUUAUAUACUAAGCAACUAGUUUCGUUUAUCAUAAGAUUAUGGUAAUCGAAACUAAAG- .........((((....)))).......((((..((---------((.............))))...))))......((((((((.((((((((....)))))))).))))))))...- ( -21.82, z-score = -2.26, R) >droSec1.super_1 7495864 115 + 14215200 AAUAUAAAUGCGUAUAAGCCCAUAUUCAUGGCUUUACUUAA---AGGCUUUGUUUUAUAACUUUUAGUCUAAGCAACUAGUUUUGCUUAGCAUAAGAUUAUGGUAAUCGAAGCUAAAG- ...........(((.(((((.........))))))))....---.(((((((..((((((.(((((..((((((((......))))))))..)))))))))))....)))))))....- ( -27.70, z-score = -2.00, R) >droSim1.chr2R 8409316 94 + 19596830 ---------------------AAUAUGACAGCUUCACUUAA---AGGCUUUAUUUUAUAUCUUUUAUUCUGAGCAACUAGUUUUGCUUAGCAUAAGAUUAUGGUAAUCGAAGCUAAAG- ---------------------........((((((...(((---((......)))))((((((((((.((((((((......)))))))).))))))....))))...))))))....- ( -20.80, z-score = -1.52, R) >consensus ____________UAUUAGCACAUAUUCAUAGCUUCACUUAA___AGGCUUUGUUUUAUAUCUUUUAUUCUAAGCAACUAGUUUUGCUUAGCAUAAGAUUAUGGUAAUCGAAGCUAAAG_ ..............................................((((....................))))...((((((((.(((.((((....)))).))).)))))))).... ( -9.26 = -8.98 + -0.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:21:47 2011