| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,949,396 – 9,949,486 |
| Length | 90 |
| Max. P | 0.999842 |

| Location | 9,949,396 – 9,949,486 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 60.64 |
| Shannon entropy | 0.79529 |
| G+C content | 0.40579 |
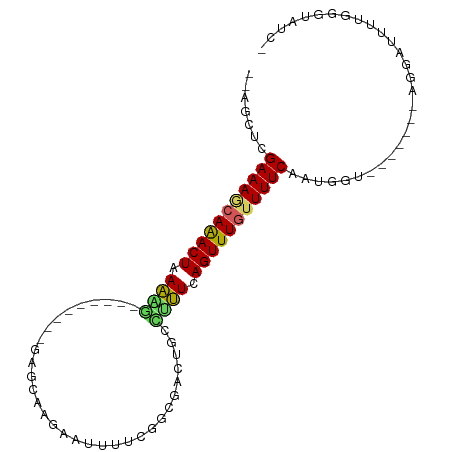
| Mean single sequence MFE | -22.63 |
| Consensus MFE | -11.67 |
| Energy contribution | -11.44 |
| Covariance contribution | -0.23 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.59 |
| SVM RNA-class probability | 0.999000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
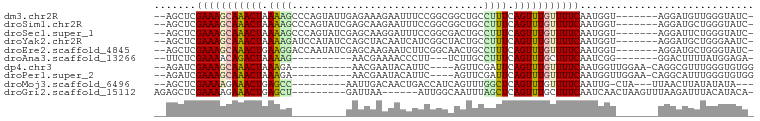
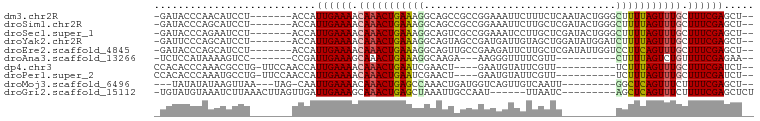
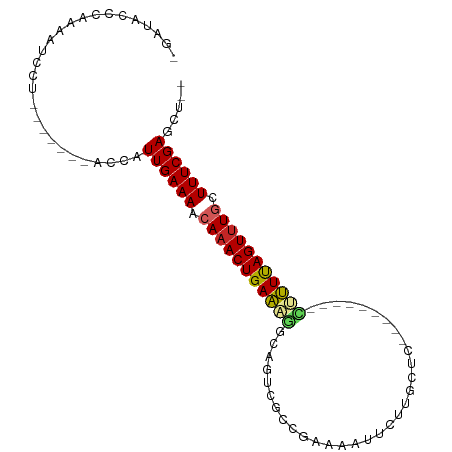
>dm3.chr2R 9949396 90 + 21146708 --AGCUCGAAAGCAAACUAAAAGCCCAGUAUUGAGAAAGAAUUUCCGGCGGCUGCCUUUCAGUUUGUUUUCAAUGGU-------AGGAUGUUGGGUAUC- --.(((((((((((((((.((((..((((.(((.(((.....))))))..)))).)))).)))))))))))((((..-------....))))))))...- ( -21.90, z-score = -0.41, R) >droSim1.chr2R 8386594 90 + 19596830 --AGCUCGAAAGCAAACUAAAAGCCCAGUAUCGAGCAAGAAUUUCCGGCGGCUGCCUUUCAGUUUGUUUUCAAUGGU-------AGGAUGCUGGGUAUC- --.(((....))).........((((((((((..((..(.....)..))..(((((..................)))-------))))))))))))...- ( -26.77, z-score = -1.38, R) >droSec1.super_1 7473863 90 + 14215200 --AGCUCGAAAGCAAACUAAAAGCCCAGUAUCGAGCAAGGAUUUCCGGCGACUGCCUUUCAGUUUGUUUUCAAUGGU-------AGGAUUCUGGGUAUC- --.(((((((((((((((.((((..((((.....((..((....)).)).)))).)))).)))))))))))...((.-------.....)).))))...- ( -24.70, z-score = -1.13, R) >droYak2.chr2R 9898198 90 + 21139217 --AGCUCGAAAGCAAACUAAAAGAUCCAUAUCCAGCUACAAUCAUCGGCUACUGCCUUUCAGUUUGUUUUCAAUGGU-------AGGAUGCUGGGAAUC- --.(((....)))...........(((((((((.((((........(((....))).......(((....)))))))-------.))))).))))....- ( -18.90, z-score = -0.24, R) >droEre2.scaffold_4845 6716969 90 - 22589142 --AGCUCGAAAGCAAACUGAAGGACCAAUAUCGAGCAAGAAUCUUCGGCAACUGCCUUUCAGUUUGUUUUCAAUGGU-------AGGAUGCUGGGUAUC- --.((((((((((((((((((((........((((........))))((....))))))))))))))))))...(((-------.....)))))))...- ( -27.80, z-score = -2.27, R) >droAna3.scaffold_13266 13933787 77 - 19884421 --UUCUCGAAAACAGACUAAAAG----------AACGAAAACCCUU---UCUUGCCUUUCAGUUUGCUUUCAAUCGG-------GGACUUUUAUGGAGA- --.((((((((.((((((.((((----------.(.((((....))---)).)..)))).)))))).))))....(.-------...).......))))- ( -17.20, z-score = -0.90, R) >dp4.chr3 2658999 83 + 19779522 --AGAUCGAAAGCAAACUAAAGA----------AACGAAUACAUUC----AGUUCGAUUCAGUUUGUUUUCAAUGGUUGGAA-CAGGCGUUUGGGUGUGG --.....(((((((((((...((----------(.(((((......----.))))).))))))))))))))...........-((.((......)).)). ( -20.70, z-score = -1.90, R) >droPer1.super_2 2846017 83 + 9036312 --AGAUCGAAAGCAAACUAAAGA----------AACGAAUACAUUC----AGUUCGAUUCAGUUUGUUUUCAAUGGUUGGAA-CAGGCAUUUGGGUGUGG --.....(((((((((((...((----------(.(((((......----.))))).))))))))))))))...........-((.((......)).)). ( -20.50, z-score = -2.10, R) >droMoj3.scaffold_6496 23294599 82 + 26866924 --AGCUCGAAAAGAAACUGAGCC---------AAUUGACAACUGACCAUCAGUUUGGCUCAGUUUGUUUUCAAUUG-CUA---UUAACUUAUAUAUA--- --(((..(((((.((((((((((---------((......((((.....)))))))))))))))).)))))....)-)).---..............--- ( -28.80, z-score = -6.20, R) >droGri2.scaffold_15112 1096647 84 - 5172618 AGAGCUCGAAAAGAAACUGAGCU---------GAUUAA------AUUGGCAAUUUAGCUCAGUUUGCUUUCAAUCAACUAAGUUUAAGAUUUACAUACA- .......((((..((((((((((---------((....------.........))))))))))))..))))............................- ( -19.02, z-score = -1.78, R) >consensus __AGCUCGAAAGCAAACUAAAAG_________GAGCAAGAAUUUUCGGCGACUGCCUUUCAGUUUGUUUUCAAUGGU_______AGGAUUUUGGGUAUC_ .......(((((((((((.((((................................)))).)))))))))))............................. (-11.67 = -11.44 + -0.23)
| Location | 9,949,396 – 9,949,486 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 60.64 |
| Shannon entropy | 0.79529 |
| G+C content | 0.40579 |
| Mean single sequence MFE | -20.86 |
| Consensus MFE | -13.92 |
| Energy contribution | -13.73 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.55 |
| SVM RNA-class probability | 0.999842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9949396 90 - 21146708 -GAUACCCAACAUCCU-------ACCAUUGAAAACAAACUGAAAGGCAGCCGCCGGAAAUUCUUUCUCAAUACUGGGCUUUUAGUUUGCUUUCGAGCU-- -...............-------....((((((.(((((((((((.(((.....((((.....)))).....)).).))))))))))).))))))...-- ( -20.20, z-score = -1.55, R) >droSim1.chr2R 8386594 90 - 19596830 -GAUACCCAGCAUCCU-------ACCAUUGAAAACAAACUGAAAGGCAGCCGCCGGAAAUUCUUGCUCGAUACUGGGCUUUUAGUUUGCUUUCGAGCU-- -...............-------....((((((.(((((((((((((....)))..........(((((....))))))))))))))).))))))...-- ( -23.50, z-score = -1.46, R) >droSec1.super_1 7473863 90 - 14215200 -GAUACCCAGAAUCCU-------ACCAUUGAAAACAAACUGAAAGGCAGUCGCCGGAAAUCCUUGCUCGAUACUGGGCUUUUAGUUUGCUUUCGAGCU-- -...............-------....((((((.(((((((((((((....)))((....))..(((((....))))))))))))))).))))))...-- ( -24.40, z-score = -2.01, R) >droYak2.chr2R 9898198 90 - 21139217 -GAUUCCCAGCAUCCU-------ACCAUUGAAAACAAACUGAAAGGCAGUAGCCGAUGAUUGUAGCUGGAUAUGGAUCUUUUAGUUUGCUUUCGAGCU-- -...............-------....((((((.(((((((((((((.(((.(((...........))).))).).)))))))))))).))))))...-- ( -20.10, z-score = -0.50, R) >droEre2.scaffold_4845 6716969 90 + 22589142 -GAUACCCAGCAUCCU-------ACCAUUGAAAACAAACUGAAAGGCAGUUGCCGAAGAUUCUUGCUCGAUAUUGGUCCUUCAGUUUGCUUUCGAGCU-- -...............-------....((((((.(((((((((.(((...((.(((..........))).))...))).))))))))).))))))...-- ( -20.70, z-score = -1.09, R) >droAna3.scaffold_13266 13933787 77 + 19884421 -UCUCCAUAAAAGUCC-------CCGAUUGAAAGCAAACUGAAAGGCAAGA---AAGGGUUUUCGUU----------CUUUUAGUCUGUUUUCGAGAA-- -((((......((((.-------..))))(((((((.(((((((((...((---((....))))..)----------)))))))).))))))))))).-- ( -20.80, z-score = -2.11, R) >dp4.chr3 2658999 83 - 19779522 CCACACCCAAACGCCUG-UUCCAACCAUUGAAAACAAACUGAAUCGAACU----GAAUGUAUUCGUU----------UCUUUAGUUUGCUUUCGAUCU-- .................-........(((((((.(((((((((..(((.(----(((....)))).)----------))))))))))).)))))))..-- ( -15.90, z-score = -2.90, R) >droPer1.super_2 2846017 83 - 9036312 CCACACCCAAAUGCCUG-UUCCAACCAUUGAAAACAAACUGAAUCGAACU----GAAUGUAUUCGUU----------UCUUUAGUUUGCUUUCGAUCU-- .................-........(((((((.(((((((((..(((.(----(((....)))).)----------))))))))))).)))))))..-- ( -15.90, z-score = -2.95, R) >droMoj3.scaffold_6496 23294599 82 - 26866924 ---UAUAUAUAAGUUAA---UAG-CAAUUGAAAACAAACUGAGCCAAACUGAUGGUCAGUUGUCAAUU---------GGCUCAGUUUCUUUUCGAGCU-- ---..............---.((-(..(((((((.((((((((((((..(((..(....)..))).))---------)))))))))).))))))))))-- ( -28.50, z-score = -5.47, R) >droGri2.scaffold_15112 1096647 84 + 5172618 -UGUAUGUAAAUCUUAAACUUAGUUGAUUGAAAGCAAACUGAGCUAAAUUGCCAAU------UUAAUC---------AGCUCAGUUUCUUUUCGAGCUCU -..........................(((((((.((((((((((..((((.....------.)))).---------)))))))))).)))))))..... ( -18.60, z-score = -1.99, R) >consensus _GAUACCCAAAAUCCU_______ACCAUUGAAAACAAACUGAAAGGCAGUCGCCGAAAAUUCUUGCUC_________CUUUUAGUUUGCUUUCGAGCU__ ...........................((((((.(((((((((((................................))))))))))).))))))..... (-13.92 = -13.73 + -0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:21:43 2011