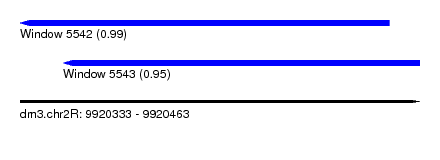
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,920,333 – 9,920,463 |
| Length | 130 |
| Max. P | 0.987193 |

| Location | 9,920,333 – 9,920,453 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.79 |
| Shannon entropy | 0.34307 |
| G+C content | 0.54438 |
| Mean single sequence MFE | -25.28 |
| Consensus MFE | -22.74 |
| Energy contribution | -23.14 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.987193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9920333 120 - 21146708 CUACUUCAUGGCAUUGUGAGCAUGCCCCACUUUUUGGUGAGGGCAUAACAAACGACCAAUUGCUCGCUCAGAUCUGCUCCUACCCUCUCCGCUGCCACUAACCCCCCCUCCCCCCUGCAC ........(((((..((((((((((((((((....)))).))))))..(....).......))))))........((.............)))))))....................... ( -28.42, z-score = -2.21, R) >droSim1.chr2R_random 2322596 102 - 2996586 CUACUUCAU-GCA-UGUGAGCAUG-CCCACUUUU--GUGAGGGCAU-ACAAACGACCAAUUGCUCGCUCAGAUCUGCUCCU-CCCUAA---------CUAACCCCCCUCCCUCUCCGC-- .........-(((-(.((((((((-(((.(....--..).))))))-......((.(....).))))))).)..)))....-......---------.....................-- ( -20.10, z-score = -1.65, R) >droSec1.super_1 7445002 108 - 14215200 CUACUUCAUGGCAUUGUGAGCAUGCCCCACUUUUUGGUGAGGGCAUAACAACCGACCAAUUGCUCGCUCAGAUCUGCUCCU-CCCUAU---------CUAAUCCCCCUCCCUCUCCGC-- .........((((.(.(((((((((((((((....)))).)))))).......((.(....).))))))).)..))))...-......---------.....................-- ( -26.30, z-score = -2.17, R) >droYak2.chr2R 14106508 111 + 21139217 CUACUUCAUGGCAUUGUGAGCAUGCCCCACUUUUCGGUGAGGGCAUAACAAACGACCAAUUGCUCGCUCAGAUCUGCACUUCCUCCUCCCCC--ACCUCAACCCCCCAUUUCC------- ..........(((.(.(((((((((((((((....)))).)))))).......((.(....).))))))).)..)))...............--...................------- ( -25.40, z-score = -2.25, R) >droEre2.scaffold_4845 6688330 117 + 22589142 CUACUUCAUGGCAUUGUGAGCAUGCCCCACUUUUUGGUGAGGGCAUAACAAACGACCAAUUGCUCGCUCAGAUCUGCACUUCCUCCUGCUCC--ACCCCAACCCCCCUCCCUCAUUUCC- ........(((...((.((((((((((((((....)))).))))))...................((........))..........)))))--)..)))...................- ( -26.20, z-score = -1.94, R) >consensus CUACUUCAUGGCAUUGUGAGCAUGCCCCACUUUUUGGUGAGGGCAUAACAAACGACCAAUUGCUCGCUCAGAUCUGCUCCU_CCCUAUC__C___C_CUAACCCCCCUCCCUCUCCGC__ ..........(((.(.(((((((((((((((....)))).)))))).......((.(....).))))))).)..)))........................................... (-22.74 = -23.14 + 0.40)

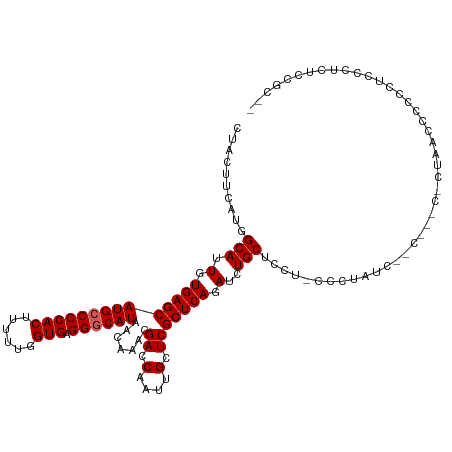
| Location | 9,920,347 – 9,920,463 |
|---|---|
| Length | 116 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.83 |
| Shannon entropy | 0.45352 |
| G+C content | 0.52192 |
| Mean single sequence MFE | -26.53 |
| Consensus MFE | -22.38 |
| Energy contribution | -22.24 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9920347 116 - 21146708 ----CAAACGUGUUCUACUUCAUGGCAUUGUGAGCAUGCCCCACUUUUUGGUGAGGGCAUAACAAACGACCAAUUGCUCGCUCAGAUCUGCUCCUACCCUCUCCGCUGCCACUAACCCCC ----..................(((((..((((((((((((((((....)))).))))))..(....).......))))))........((.............)))))))......... ( -28.42, z-score = -1.60, R) >droSim1.chr2R_random 2322608 99 - 2996586 ----CCAACGUG-UCUACUUCAU-GCA-UGUGAGCAUG-CCCACUUUU--GUGAGGGCAU-ACAAACGACCAAUUGCUCGCUCAGAUCUGCUCCUCCCUAACUAACCCCC---------- ----...(((((-(.........-)))-)))(((((((-(((.(....--..).))))))-.....(((.(....).))).........)))).................---------- ( -20.20, z-score = -1.04, R) >droSec1.super_1 7445014 106 - 14215200 ----CAAACGUGUUCUACUUCAUGGCAUUGUGAGCAUGCCCCACUUUUUGGUGAGGGCAUAACAACCGACCAAUUGCUCGCUCAGAUCUGCUCCUCCCUAUCUAAUCCCC---------- ----....((((........))))(((.(.(((((((((((((((....)))).)))))).......((.(....).))))))).)..)))...................---------- ( -28.20, z-score = -2.35, R) >droYak2.chr2R 14106515 114 + 21139217 ----CAAACGUGUUCUACUUCAUGGCAUUGUGAGCAUGCCCCACUUUUCGGUGAGGGCAUAACAAACGACCAAUUGCUCGCUCAGAUCUGCACUU-CCUCCUCCCCCACC-UCAACCCCC ----....((((........))))(((.(.(((((((((((((((....)))).)))))).......((.(....).))))))).)..)))....-..............-......... ( -28.00, z-score = -2.27, R) >droEre2.scaffold_4845 6688343 114 + 22589142 ----CAAACGUGUUCUACUUCAUGGCAUUGUGAGCAUGCCCCACUUUUUGGUGAGGGCAUAACAAACGACCAAUUGCUCGCUCAGAUCUGCACUU-CCUCCUGCUCCACC-CCAACCCCC ----....((((........))))(((.(.(((((((((((((((....)))).)))))).......((.(....).))))))).)..)))....-..............-......... ( -28.70, z-score = -1.89, R) >dp4.chr3 8564049 98 + 19779522 CAAAACUGUGUGCUCUACUUCAUGGCACUGUGAGCAUGCCCCACAUUU-GGUGAGGGCAUAGCAAACGACCAAUUUCUCGCUCGGAUCUGUGGCCUCCU--------------------- .......((((((((.((...........)))))))))).(((....)-)).((((.(((((.(..(((.(........).)))..)))))).))))..--------------------- ( -26.60, z-score = -0.07, R) >droPer1.super_4 542863 98 + 7162766 CAAAACUGUGCGCUCUACUUCAUGGCACUGUGAGCAUGCCCCACAUUU-GGUGAGGGCAUAGCAAACGACCAAUUUCUCGCUCGGAUCUGUGGCCUCCU--------------------- ........(((((((.((...........))))))(((((((((....-.))).)))))).)))...(.(((...(((.....)))....))).)....--------------------- ( -25.60, z-score = 0.27, R) >consensus ____CAAACGUGUUCUACUUCAUGGCAUUGUGAGCAUGCCCCACUUUU_GGUGAGGGCAUAACAAACGACCAAUUGCUCGCUCAGAUCUGCUCCU_CCU_CU_C_CC_CC__________ ........((((........))))(((.(.((((((((((((((......))).)))))).......((........))))))).)..)))............................. (-22.38 = -22.24 + -0.14)
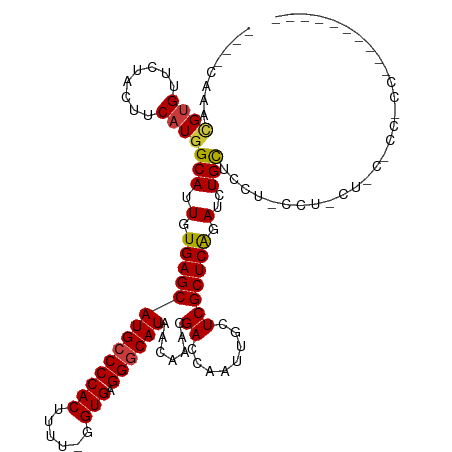
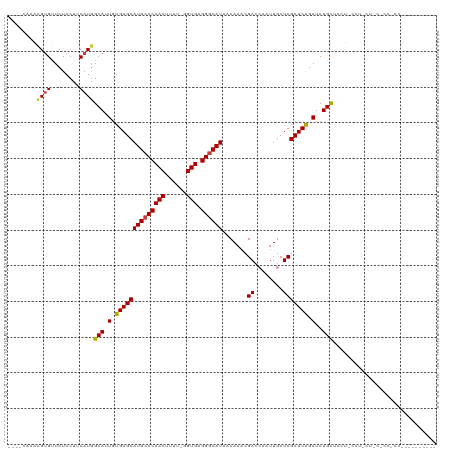
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:21:42 2011