| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,903,915 – 9,904,040 |
| Length | 125 |
| Max. P | 0.705511 |

| Location | 9,903,915 – 9,904,010 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 89.39 |
| Shannon entropy | 0.19309 |
| G+C content | 0.53387 |
| Mean single sequence MFE | -36.87 |
| Consensus MFE | -29.59 |
| Energy contribution | -31.07 |
| Covariance contribution | 1.47 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.606963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
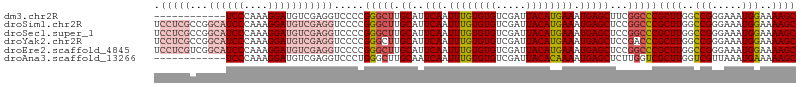
>dm3.chr2R 9903915 95 + 21146708 ------------UCCCAAAGGAUGUCGAGGUCCCCGGGCUUGCAUUCAAUUUGUGUGUCGAUUACAUGAAAUGAGCUUCGGCCCGCUUGGCCGGGAAAUGGAAAAGC ------------..(((..((((......))))((((.(..((..(((.((..((((.....))))..)).)))((....))..))..).))))....)))...... ( -30.80, z-score = -0.73, R) >droSim1.chr2R 8340467 107 + 19596830 UCCUCGCCGGCAUCCCAAAGGAUGUCGAGGUCCCCGGGCUUGCAUUCAAUUUGUGUGUCGAUUACAUGAAAUGAGCUCCGGCCCGCUUGGCCGGGAAAUGGAAAAGC (((...(((((((((....)))))))..((((..((((((.(..((((.((..((((.....))))..)).))))..).))))))...)))).))....)))..... ( -42.00, z-score = -2.03, R) >droSec1.super_1 7428946 107 + 14215200 UCCUCGCCGGCAUCCCAAAGGAUGUCGAGGUCCCCGGGCUUGCAUUCAAUUUGUGUGUCGAUUACAUGAAAUGAGCUCCGGCCCGCUUGGCCGGGAAAUGGAAAAGC (((...(((((((((....)))))))..((((..((((((.(..((((.((..((((.....))))..)).))))..).))))))...)))).))....)))..... ( -42.00, z-score = -2.03, R) >droYak2.chr2R 14089840 107 - 21139217 UCCUCGCCGGCAUCCCAAAGGAUGUCGAGGUCCCCGGGCUUGCAUUCAAUUUGUGUGUCGAUUACAUGAAAUGAGCUCCGACCCGCUUGGCCGGGAAAUGGAAAAGC (((...(((((((((....)))))))..((((..((((.(((..((((.((..((((.....))))..)).))))...)))))))...)))).))....)))..... ( -37.40, z-score = -1.16, R) >droEre2.scaffold_4845 6671752 107 - 22589142 UCCUCGUCGGCAUCCCAAAGGAUGUCGAGGUCCCCGGGCUUGCAUUCAAUUUGUGUGUCGAUUACAUGAAAUGAGCUCCGGCCCGCUUGGCCGGGAAAUGGAAAAGC ((((..(((((((((....)))))))))((((..((((((.(..((((.((..((((.....))))..)).))))..).))))))...))))))))........... ( -43.20, z-score = -2.59, R) >droAna3.scaffold_13266 13888531 95 - 19884421 ------------UCCCAAAGGAUGUCGAGGUCCCUGGGCUUGCAAUCAAUUUGUGUGUCGAUUACACAAAAUGAGCUCUUGGUCGCUUGGUCGUUAAAUGAAAAAGC ------------..((((..(..(((.((....)).)))......(((.((((((((.....)))))))).))).)..))))..((((..((((...))))..)))) ( -25.80, z-score = -1.36, R) >consensus UCCUCGCCGGCAUCCCAAAGGAUGUCGAGGUCCCCGGGCUUGCAUUCAAUUUGUGUGUCGAUUACAUGAAAUGAGCUCCGGCCCGCUUGGCCGGGAAAUGGAAAAGC .......((((((((....))))))))........(((((.((..(((.((((((((.....)))))))).)))))...)))))((((..(((.....)))..)))) (-29.59 = -31.07 + 1.47)

| Location | 9,903,943 – 9,904,040 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 78.90 |
| Shannon entropy | 0.39754 |
| G+C content | 0.42989 |
| Mean single sequence MFE | -27.66 |
| Consensus MFE | -19.48 |
| Energy contribution | -18.23 |
| Covariance contribution | -1.26 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.705511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9903943 97 + 21146708 UGCAUUCAAUUUGUGUGUCGAUUACAUGAAAUGAG-CUUCGGCCC---GCUUGGCCGGGAAAUGGAAAAGCUGAUGAAGAGCGGGUUU---------UAUAUGCAAAAUU- ((((((((.((..((((.....))))..)).))).-....(((((---((((..((((............)))..)..))))))))).---------...))))).....- ( -26.10, z-score = -0.86, R) >droSim1.chr2R 8340507 97 + 19596830 UGCAUUCAAUUUGUGUGUCGAUUACAUGAAAUGAG-CUCCGGCCC---GCUUGGCCGGGAAAUGGAAAAGCUGAUGAAGAGCGGGUUU---------UAUAUGCAAAAUU- ((((((((.((..((((.....))))..)).))).-.(((((((.---....)))))))..((((((..(((.......)))...)))---------)))))))).....- ( -28.90, z-score = -1.56, R) >droSec1.super_1 7428986 97 + 14215200 UGCAUUCAAUUUGUGUGUCGAUUACAUGAAAUGAG-CUCCGGCCC---GCUUGGCCGGGAAAUGGAAAAGCUGAUGAAGAGCGGGUUU---------UAUAUGCAAAAUU- ((((((((.((..((((.....))))..)).))).-.(((((((.---....)))))))..((((((..(((.......)))...)))---------)))))))).....- ( -28.90, z-score = -1.56, R) >droYak2.chr2R 14089880 97 - 21139217 UGCAUUCAAUUUGUGUGUCGAUUACAUGAAAUGAG-CUCCGACCC---GCUUGGCCGGGAAAUGGAAAAGCUGAUGAAGAGCGGGUUU---------UAUAUGCAAAAUU- ((((((((.((..((((.....))))..)).))).-....(((((---((((..((((............)))..)..))))))))).---------...))))).....- ( -26.60, z-score = -1.35, R) >droEre2.scaffold_4845 6671792 97 - 22589142 UGCAUUCAAUUUGUGUGUCGAUUACAUGAAAUGAG-CUCCGGCCC---GCUUGGCCGGGAAAUGGAAAAGCUGAUGAAGAGCGGGUUU---------UAUAUGCAAAAUU- ((((((((.((..((((.....))))..)).))).-.(((((((.---....)))))))..((((((..(((.......)))...)))---------)))))))).....- ( -28.90, z-score = -1.56, R) >droAna3.scaffold_13266 13888559 107 - 19884421 UGCAAUCAAUUUGUGUGUCGAUUACACAAAAUGAG-CUCUUGGUC---GCUUGGUCGUUAAAUGAAAAAGCUUCUCAUAUACGGAUAUAUCUACAUAUAUAUGCAAAAAUU ((((.(((.((((((((.....)))))))).))).-.........---((((..((((...))))..)))).............((((((....)))))).))))...... ( -21.20, z-score = -1.28, R) >droPer1.super_4 527342 109 - 7162766 UGCAUUCAAUUUGUGUAUCGAUUACACGGCAUGAGGCUCUUGUCACAGGCUGUGUGUGUGGGAGGGGGGGACCACAGAGCAUUGGAGUAU-UGCACUCAUAUGCAAAAUG- (((((.((((((.(((.(((..((((((((.((.(((....))).)).))))))))..)))....((....))))).)).))))((((..-...))))..))))).....- ( -33.00, z-score = -0.73, R) >consensus UGCAUUCAAUUUGUGUGUCGAUUACAUGAAAUGAG_CUCCGGCCC___GCUUGGCCGGGAAAUGGAAAAGCUGAUGAAGAGCGGGUUU_________UAUAUGCAAAAUU_ ((((((((.((((((((.....)))))))).)))......(((((...((((..(((.....)))..))))...........))))).............)))))...... (-19.48 = -18.23 + -1.26)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:21:39 2011