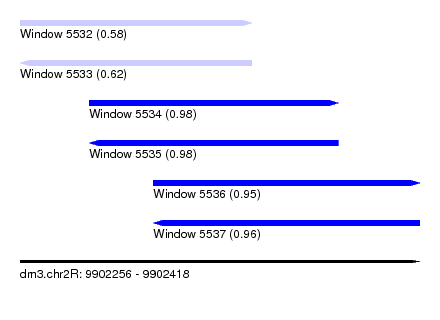
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,902,256 – 9,902,418 |
| Length | 162 |
| Max. P | 0.984509 |

| Location | 9,902,256 – 9,902,350 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 82.84 |
| Shannon entropy | 0.29710 |
| G+C content | 0.34610 |
| Mean single sequence MFE | -22.04 |
| Consensus MFE | -14.88 |
| Energy contribution | -15.28 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.579120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
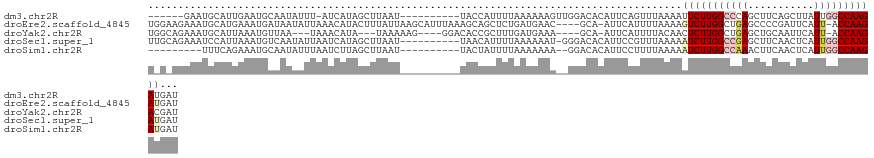
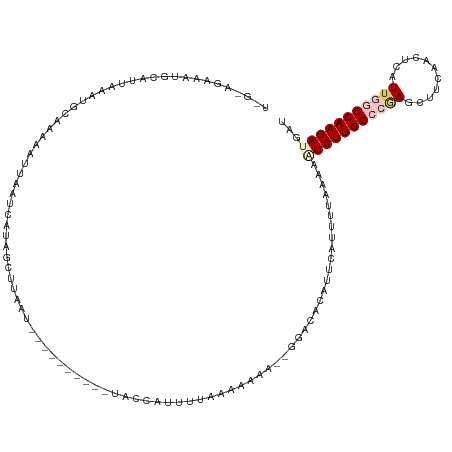
>dm3.chr2R 9902256 94 + 21146708 GAAUAAUAUUGUUGCAAAAGGCAAUUCG-----UACGAUUAUUUCUUAUCAUUUAUUGCAUCAUCUUGGCCAAUAAGCUGAAGCUGGGCCAAGAAUUUU (((((((...(((((.....)))))...-----....)))))))...................((((((((....(((....))).))))))))..... ( -23.60, z-score = -2.12, R) >droEre2.scaffold_4845 6670082 98 - 22589142 GCAUAAUAUUGUUGCAAAAGGCAAUGCAAUUUCUUCGAUUAUUUCUUAUCAUUUAUUGCAUCAUCUUGGU-AAUGAAUCGGGGCUCAGCCAAGACUUUU (((.........)))(((((...(((((((......(((........)))....)))))))..(((((((-..(((........))))))))))))))) ( -18.70, z-score = -0.48, R) >droYak2.chr2R 14088178 94 - 21139217 GCAUAAUAU-GUUGCAAAAGGCAAUGCAAAUUCUUCAAU---UCCUUAUCAUUUAUUGCAUCGUCUUGGU-AAUGAAUUGCAGCUCAGCCAAGAGUUGU .........-(((((..(((((.((((((..........---.............)))))).))))).))-))).....(((((((......))))))) ( -20.10, z-score = -0.58, R) >droSec1.super_1 7427254 91 + 14215200 GAAUAAUAUUGUUGCAAAAGGCAAUUCG-----UACGAUUA---CUUAUCAUUUAUUGCAUCAUCUUGGCCAAUGAGUUGAAGCUCGGCCAAGAUUUUU .........((.(((((...((.....)-----)..(((..---...))).....))))).))((((((((...((((....))))))))))))..... ( -22.30, z-score = -2.07, R) >droSim1.chr2R 8338828 94 + 19596830 GAAUAAUAUUGUUGCAAAAGGCAAUUCG-----UACGAUUAUUUCUUAUCGUUUAUUGCAUCAUCUUGGCCAAUGAGUUGAAGUUUGGCCAAGAUUUUU ....................(((((...-----.(((((........)))))..)))))...(((((((((((...........))))))))))).... ( -25.50, z-score = -3.09, R) >consensus GAAUAAUAUUGUUGCAAAAGGCAAUUCG_____UACGAUUAUUUCUUAUCAUUUAUUGCAUCAUCUUGGCCAAUGAGUUGAAGCUCGGCCAAGAUUUUU ..........(((((.....)))))......................................((((((((...((((....))))))))))))..... (-14.88 = -15.28 + 0.40)
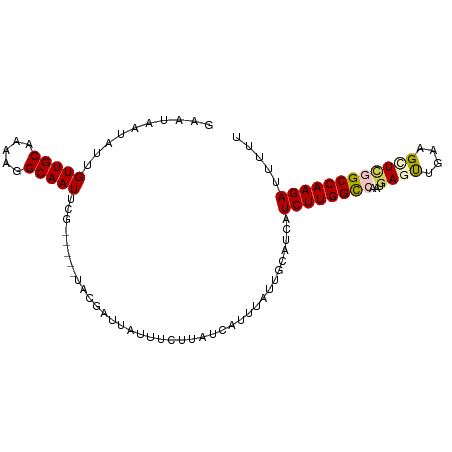
| Location | 9,902,256 – 9,902,350 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 82.84 |
| Shannon entropy | 0.29710 |
| G+C content | 0.34610 |
| Mean single sequence MFE | -19.80 |
| Consensus MFE | -12.77 |
| Energy contribution | -14.05 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.615838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9902256 94 - 21146708 AAAAUUCUUGGCCCAGCUUCAGCUUAUUGGCCAAGAUGAUGCAAUAAAUGAUAAGAAAUAAUCGUA-----CGAAUUGCCUUUUGCAACAAUAUUAUUC .....((((((((.(((....)))....))))))))....(((((..(((((.(....).))))).-----...))))).................... ( -21.40, z-score = -2.10, R) >droEre2.scaffold_4845 6670082 98 + 22589142 AAAAGUCUUGGCUGAGCCCCGAUUCAUU-ACCAAGAUGAUGCAAUAAAUGAUAAGAAAUAAUCGAAGAAAUUGCAUUGCCUUUUGCAACAAUAUUAUGC ....(((((((.((((......))))..-.)))))))((((((((...((((.(....).)))).....))))))))((.....))............. ( -17.80, z-score = -1.08, R) >droYak2.chr2R 14088178 94 + 21139217 ACAACUCUUGGCUGAGCUGCAAUUCAUU-ACCAAGACGAUGCAAUAAAUGAUAAGGA---AUUGAAGAAUUUGCAUUGCCUUUUGCAAC-AUAUUAUGC .....((((((.((((......))))..-.))))))((((((((.............---..........))))))))......(((..-......))) ( -14.40, z-score = 0.81, R) >droSec1.super_1 7427254 91 - 14215200 AAAAAUCUUGGCCGAGCUUCAACUCAUUGGCCAAGAUGAUGCAAUAAAUGAUAAG---UAAUCGUA-----CGAAUUGCCUUUUGCAACAAUAUUAUUC ....(((((((((((...........)))))))))))..(((((.....(((..(---((....))-----)..))).....)))))............ ( -21.80, z-score = -2.62, R) >droSim1.chr2R 8338828 94 - 19596830 AAAAAUCUUGGCCAAACUUCAACUCAUUGGCCAAGAUGAUGCAAUAAACGAUAAGAAAUAAUCGUA-----CGAAUUGCCUUUUGCAACAAUAUUAUUC ....(((((((((((...........)))))))))))...(((((..(((((.(....).))))).-----...))))).................... ( -23.60, z-score = -4.31, R) >consensus AAAAAUCUUGGCCGAGCUUCAACUCAUUGGCCAAGAUGAUGCAAUAAAUGAUAAGAAAUAAUCGUA_____CGAAUUGCCUUUUGCAACAAUAUUAUUC ....(((((((((((...........)))))))))))..(((((...(((((........))))).................)))))............ (-12.77 = -14.05 + 1.28)
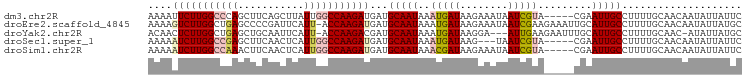
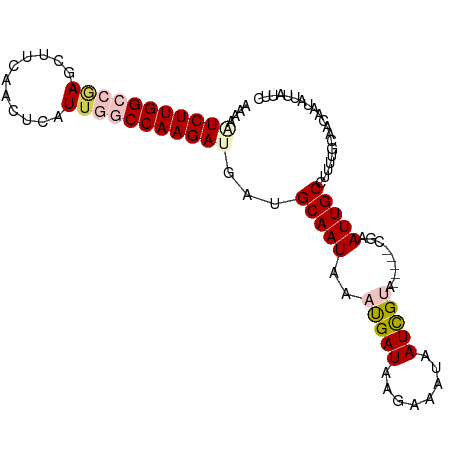
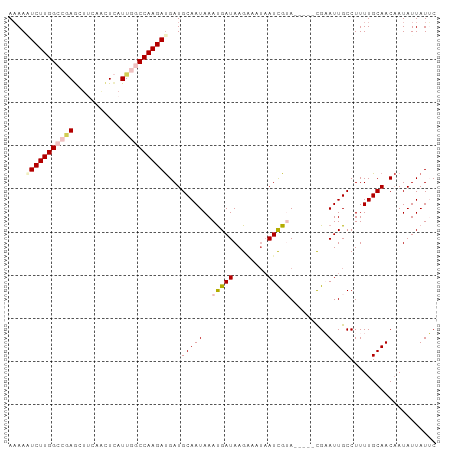
| Location | 9,902,284 – 9,902,385 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 71.66 |
| Shannon entropy | 0.48194 |
| G+C content | 0.33249 |
| Mean single sequence MFE | -23.10 |
| Consensus MFE | -12.50 |
| Energy contribution | -13.94 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.975943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9902284 101 + 21146708 -UACGAUUAUUUCUUAUCAUUUAUUGCAUCAUCUUGGCCAAUAAGCUGAAGCUGGGCCAAGAAUUUUAAACUGAAUGUGUCCAACUUUUUUAAAAUGGUAAU---------- -............(((((((((.........((((((((....(((....))).)))))))).........((((.((.....))...))))))))))))).---------- ( -23.70, z-score = -2.39, R) >droEre2.scaffold_4845 6670114 106 - 22589142 CUUCGAUUAUUUCUUAUCAUUUAUUGCAUCAUCUUGGU-AAUGAAUCGGGGCUCAGCCAAGACUUUUAAAAUGAAU-UGCG----UUCAUCAGAGCUGCUUUAAAUGCUUAA .................((((((..(((...(((((((-..(((........))))))))))..............-...(----(((....)))))))..))))))..... ( -19.90, z-score = -0.12, R) >droYak2.chr2R 14088209 99 - 21139217 CUUCAA---UUCCUUAUCAUUUAUUGCAUCGUCUUGGU-AAUGAAUUGCAGCUCAGCCAAGAGUUGUAAAAUGAAU-UGCU----UUCAUCAAAGCGGUGUCCCUUUU---- ......---................((((((..(((((-......(((((((((......)))))))))...(((.-....----))))))))..)))))).......---- ( -24.00, z-score = -2.56, R) >droSec1.super_1 7427282 97 + 14215200 -UACGAUUA---CUUAUCAUUUAUUGCAUCAUCUUGGCCAAUGAGUUGAAGCUCGGCCAAGAUUUUUAAACGGAAUGUGUCCCA-UUUUUUAAAAUGUUAAU---------- -...(((..---...))).......(((..(((((((((...((((....)))))))))))))(((((((..(((((.....))-)))))))))))))....---------- ( -21.30, z-score = -1.90, R) >droSim1.chr2R 8338856 99 + 19596830 -UACGAUUAUUUCUUAUCGUUUAUUGCAUCAUCUUGGCCAAUGAGUUGAAGUUUGGCCAAGAUUUUUAAAAGGAAUGUGUCCU--UUUUUUAAAAUAGUAAU---------- -.(((((........)))))..(((((....((((((((((...........))))))))))((((.(((((((.....))))--)))...))))..)))))---------- ( -26.60, z-score = -3.91, R) >consensus _UACGAUUAUUUCUUAUCAUUUAUUGCAUCAUCUUGGCCAAUGAGUUGAAGCUCGGCCAAGAUUUUUAAAAUGAAUGUGUCC___UUUUUUAAAAUGGUAAU__________ .................((((((((......((((((((...((((....)))))))))))).......))))))))................................... (-12.50 = -13.94 + 1.44)


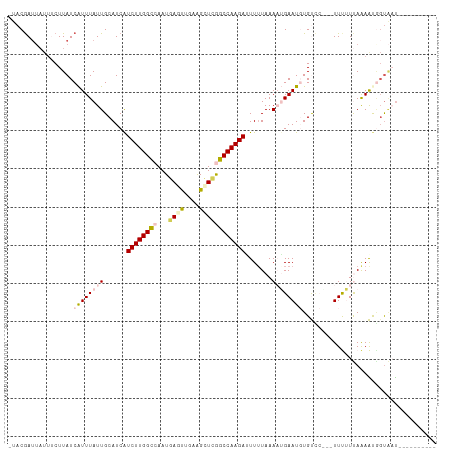
| Location | 9,902,284 – 9,902,385 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 71.66 |
| Shannon entropy | 0.48194 |
| G+C content | 0.33249 |
| Mean single sequence MFE | -21.56 |
| Consensus MFE | -11.42 |
| Energy contribution | -13.42 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.984509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9902284 101 - 21146708 ----------AUUACCAUUUUAAAAAAGUUGGACACAUUCAGUUUAAAAUUCUUGGCCCAGCUUCAGCUUAUUGGCCAAGAUGAUGCAAUAAAUGAUAAGAAAUAAUCGUA- ----------......((((((((.....((....)).....))))))))((((((((.(((....)))....))))))))...........(((((.(....).))))).- ( -21.60, z-score = -1.89, R) >droEre2.scaffold_4845 6670114 106 + 22589142 UUAAGCAUUUAAAGCAGCUCUGAUGAA----CGCA-AUUCAUUUUAAAAGUCUUGGCUGAGCCCCGAUUCAUU-ACCAAGAUGAUGCAAUAAAUGAUAAGAAAUAAUCGAAG .....((((((..(((.....((((((----....-.))))))......(((((((.((((......))))..-.)))))))..)))..))))))................. ( -18.30, z-score = -0.45, R) >droYak2.chr2R 14088209 99 + 21139217 ----AAAAGGGACACCGCUUUGAUGAA----AGCA-AUUCAUUUUACAACUCUUGGCUGAGCUGCAAUUCAUU-ACCAAGACGAUGCAAUAAAUGAUAAGGAA---UUGAAG ----..........(((((((....))----))).-..((((((..((.(((((((.((((......))))..-.)))))).).))....))))))...))..---...... ( -17.80, z-score = -0.54, R) >droSec1.super_1 7427282 97 - 14215200 ----------AUUAACAUUUUAAAAAA-UGGGACACAUUCCGUUUAAAAAUCUUGGCCGAGCUUCAACUCAUUGGCCAAGAUGAUGCAAUAAAUGAUAAG---UAAUCGUA- ----------.....((((.....(((-(((((....))))))))....(((((((((((...........)))))))))))))))......(((((...---..))))).- ( -24.30, z-score = -3.18, R) >droSim1.chr2R 8338856 99 - 19596830 ----------AUUACUAUUUUAAAAAA--AGGACACAUUCCUUUUAAAAAUCUUGGCCAAACUUCAACUCAUUGGCCAAGAUGAUGCAAUAAACGAUAAGAAAUAAUCGUA- ----------..............(((--((((.....)))))))....(((((((((((...........)))))))))))..........(((((.(....).))))).- ( -25.80, z-score = -5.70, R) >consensus __________AUUACCAUUUUAAAAAA___GGACACAUUCAUUUUAAAAAUCUUGGCCGAGCUUCAACUCAUUGGCCAAGAUGAUGCAAUAAAUGAUAAGAAAUAAUCGUA_ ......................................((((((.....(((((((((((...........)))))))))))........))))))................ (-11.42 = -13.42 + 2.00)


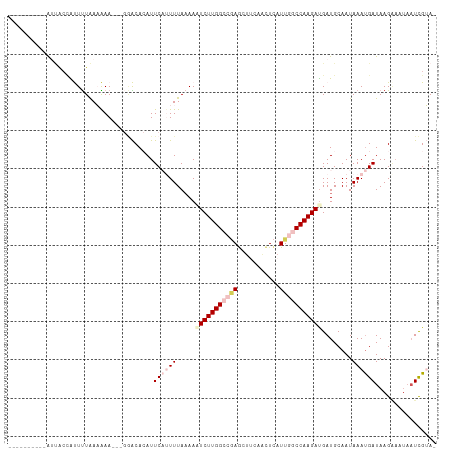
| Location | 9,902,310 – 9,902,418 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 125 |
| Reading direction | forward |
| Mean pairwise identity | 65.77 |
| Shannon entropy | 0.58234 |
| G+C content | 0.31901 |
| Mean single sequence MFE | -25.56 |
| Consensus MFE | -11.28 |
| Energy contribution | -11.68 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.950747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
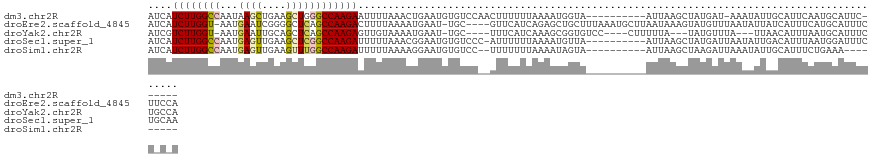
>dm3.chr2R 9902310 108 + 21146708 AUCAUCUUGGCCAAUAAGCUGAAGCUGGGCCAAGAAUUUUAAACUGAAUGUGUCCAACUUUUUUAAAAUGGUA----------AUUAAGCUAUGAU-AAAUAUUGCAUUCAAUGCAUUC------ ....((((((((....(((....))).)))))))).........((((((((.........((((..(((((.----------.....)))))..)-)))...))))))))........------ ( -26.60, z-score = -2.27, R) >droEre2.scaffold_4845 6670141 119 - 22589142 AUCAUCUUGGU-AAUGAAUCGGGGCUCAGCCAAGACUUUUAAAAUGAAU-UGC----GUUCAUCAGAGCUGCUUUAAAUGCUUAAUAAAGUAUGUUUAAUAUUAUCAUUUCAUGCAUUUCUUCCA ....(((((((-..(((........))))))))))..........(((.-(((----((((....))))........((((((....))))))....................))).)))..... ( -21.00, z-score = 0.09, R) >droYak2.chr2R 14088233 109 - 21139217 AUCGUCUUGGU-AAUGAAUUGCAGCUCAGCCAAGAGUUGUAAAAUGAAU-UGC----UUUCAUCAAAGCGGUGUCC----CUUUUUA---UAUGUUUA---UUAACAUUUAAUGCAUUUCUGCCA .......((((-(.....(((((((((......)))))))))...(((.-(((----...((((.....))))...----....(((---.(((((..---..))))).))).))).)))))))) ( -24.70, z-score = -1.66, R) >droSec1.super_1 7427305 114 + 14215200 AUCAUCUUGGCCAAUGAGUUGAAGCUCGGCCAAGAUUUUUAAACGGAAUGUGUCCC-AUUUUUUAAAAUGUUA----------AUUAAGCUAUGAUUAAUAUUGACAUUUAAUGGAUUUCUGCAA ...(((((((((...((((....))))))))))))).......(((((...((((.-(((......(((((((----------((((.....))))))))))).......))))))))))))... ( -30.92, z-score = -3.30, R) >droSim1.chr2R 8338882 104 + 19596830 AUCAUCUUGGCCAAUGAGUUGAAGUUUGGCCAAGAUUUUUAAAAGGAAUGUGUCC--UUUUUUUAAAAUAGUA----------AUUAAGCUAAGAUUAAAUAUUGCAUUUCUGAAA--------- .(((((((((((((...........))))))))))).......(((((((((...--....(((((..((((.----------.....))))...)))))...)))))))))))..--------- ( -24.60, z-score = -2.47, R) >consensus AUCAUCUUGGCCAAUGAGUUGAAGCUCGGCCAAGAUUUUUAAAAUGAAUGUGUCC__UUUUUUUAAAAUGGUA__________AUUAAGCUAUGAUUAAAAUUAACAUUUAAUGCAUUUCU_C_A ....((((((((...((((....)))))))))))).......................................................................................... (-11.28 = -11.68 + 0.40)
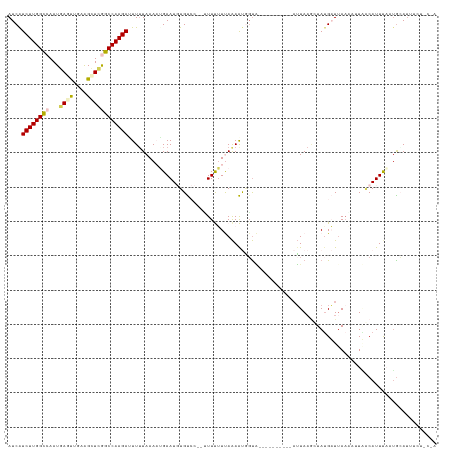
| Location | 9,902,310 – 9,902,418 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 125 |
| Reading direction | reverse |
| Mean pairwise identity | 65.77 |
| Shannon entropy | 0.58234 |
| G+C content | 0.31901 |
| Mean single sequence MFE | -22.16 |
| Consensus MFE | -10.44 |
| Energy contribution | -11.64 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9902310 108 - 21146708 ------GAAUGCAUUGAAUGCAAUAUUU-AUCAUAGCUUAAU----------UACCAUUUUAAAAAAGUUGGACACAUUCAGUUUAAAAUUCUUGGCCCAGCUUCAGCUUAUUGGCCAAGAUGAU ------...(((((...)))))......-((((.........----------....((((((((.....((....)).....))))))))((((((((.(((....)))....)))))))))))) ( -23.20, z-score = -1.40, R) >droEre2.scaffold_4845 6670141 119 + 22589142 UGGAAGAAAUGCAUGAAAUGAUAAUAUUAAACAUACUUUAUUAAGCAUUUAAAGCAGCUCUGAUGAAC----GCA-AUUCAUUUUAAAAGUCUUGGCUGAGCCCCGAUUCAUU-ACCAAGAUGAU ....(((..(((...(((((.(((((............)))))..)))))...)))..)))((((((.----...-.))))))......(((((((.((((......))))..-.)))))))... ( -20.30, z-score = -0.33, R) >droYak2.chr2R 14088233 109 + 21139217 UGGCAGAAAUGCAUUAAAUGUUAA---UAAACAUA---UAAAAAG----GGACACCGCUUUGAUGAAA----GCA-AUUCAUUUUACAACUCUUGGCUGAGCUGCAAUUCAUU-ACCAAGACGAU ..(((....))).(((.(((((..---..))))).---)))...(----(....))(((((....)))----)).-..............((((((.((((......))))..-.)))))).... ( -20.70, z-score = -1.02, R) >droSec1.super_1 7427305 114 - 14215200 UUGCAGAAAUCCAUUAAAUGUCAAUAUUAAUCAUAGCUUAAU----------UAACAUUUUAAAAAAU-GGGACACAUUCCGUUUAAAAAUCUUGGCCGAGCUUCAACUCAUUGGCCAAGAUGAU ...............((((((....(((((.......)))))----------..))))))....((((-((((....))))))))....(((((((((((...........)))))))))))... ( -23.90, z-score = -2.28, R) >droSim1.chr2R 8338882 104 - 19596830 ---------UUUCAGAAAUGCAAUAUUUAAUCUUAGCUUAAU----------UACUAUUUUAAAAAAA--GGACACAUUCCUUUUAAAAAUCUUGGCCAAACUUCAACUCAUUGGCCAAGAUGAU ---------...((....)).....(((((...(((......----------..)))..)))))((((--(((.....)))))))....(((((((((((...........)))))))))))... ( -22.70, z-score = -3.97, R) >consensus U_G_AGAAAUGCAUUAAAUGCAAAAAUUAAUCAUAGCUUAAU__________UACCAUUUUAAAAAAA__GGACACAUUCAUUUUAAAAAUCUUGGCCGAGCUUCAACUCAUUGGCCAAGAUGAU .........................................................................................(((((((((((...........)))))))))))... (-10.44 = -11.64 + 1.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:21:37 2011