| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,896,866 – 9,896,932 |
| Length | 66 |
| Max. P | 0.736273 |

| Location | 9,896,866 – 9,896,932 |
|---|---|
| Length | 66 |
| Sequences | 9 |
| Columns | 68 |
| Reading direction | reverse |
| Mean pairwise identity | 69.91 |
| Shannon entropy | 0.64649 |
| G+C content | 0.48606 |
| Mean single sequence MFE | -19.00 |
| Consensus MFE | -8.86 |
| Energy contribution | -8.81 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.43 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.736273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9896866 66 - 21146708 --AAUCAAGCAAAGCUGAUUAAAGCCAAUAAUCGCUGGCAAAACGCGAGCGGCCGUUGCGCGUUUUCU --(((((.((...)))))))...((((........))))(((((((((((....))).)))))))).. ( -19.40, z-score = -0.71, R) >droSim1.chr2R 8333423 66 - 19596830 --AAUCAAGCAAAGCUGAUUAAAGCCAAUAAUCGCUGGCAAAACGCGAGCGGCCGUUGCGCGUUUUCU --(((((.((...)))))))...((((........))))(((((((((((....))).)))))))).. ( -19.40, z-score = -0.71, R) >droSec1.super_1 7421982 66 - 14215200 --AAUCAAGCAAAGCUGAUUAAAGCCAAUAAUCGUUGGCAAAACGCGAGCGGCCGUUGCGCGUUUUCU --(((((.((...)))))))...((((((....))))))(((((((((((....))).)))))))).. ( -21.50, z-score = -1.82, R) >droYak2.chr2R 14082485 66 + 21139217 --AAUCAGGCAAAGCUGAUUAAAGCCAAUAAUCGCUGGCAAAACGCGAGCGGCCGUUGCGCGUUUUCU --((((((......))))))...((((........))))(((((((((((....))).)))))))).. ( -21.50, z-score = -1.10, R) >droEre2.scaffold_4845 6664449 66 + 22589142 --AGUCAGGCAAAGCUGAUUAAAGCCAAUAAUCGCUGGCAAAACGCGAGCGGCCGUUGCGCGUUUUCU --((((((......))))))...((((........))))(((((((((((....))).)))))))).. ( -21.50, z-score = -0.81, R) >droAna3.scaffold_13266 13881049 57 + 19884421 -----------AACAGAAGCGUAGCCACUAAUCGCUGGCAAAACGCGAGCGUUUCUAGCGCGUUUUCU -----------....(((((((.((.......((((.((.....)).))))......))))))))).. ( -16.72, z-score = -0.74, R) >dp4.chr3 8540674 64 + 19779522 ---AAGCGACAAAAACUGAUUAAAACAUUAAUCGCUAGCGAAACGCGAGCGGCUGUCGCGCGUUUCU- ---..((((((......((((((....))))))(((.(((...))).)))...))))))........- ( -19.70, z-score = -1.49, R) >droPer1.super_4 521139 64 + 7162766 ---AAGCGACAAAAACUGAUUAAAACAUUAAUCGCUAGCGAAACGCAGGCGGCUGUCGCGCGUUUCU- ---..((((((......((((((....))))))(((.(((...))).)))...))))))........- ( -19.70, z-score = -1.78, R) >droMoj3.scaffold_6496 23224643 57 - 26866924 AAUUACGCACAACUGAGAUUAAAGGCUUUAAUCGCUAGCGAGUCGCGCGCGACUUUG----------- .....(((.(....).((((((.....))))))....)))(((((....)))))...----------- ( -11.60, z-score = 0.48, R) >consensus __AAUCAGGCAAAGCUGAUUAAAGCCAAUAAUCGCUGGCAAAACGCGAGCGGCCGUUGCGCGUUUUCU ...............................(((((.((.....)).)))))................ ( -8.86 = -8.81 + -0.05)

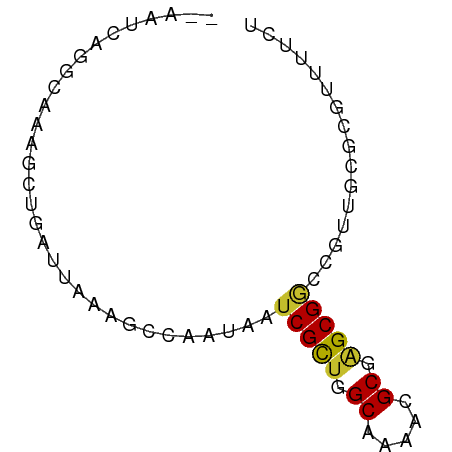

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:21:31 2011